
Bovine adenovirus 4 (BAdV-4)
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Preplasmiviricota; Tectiliviricetes; Rowavirales; Adenoviridae; Atadenovirus; Bovine atadenovirus D
Average proteome isoelectric point is 7.2
Get precalculated fractions of proteins
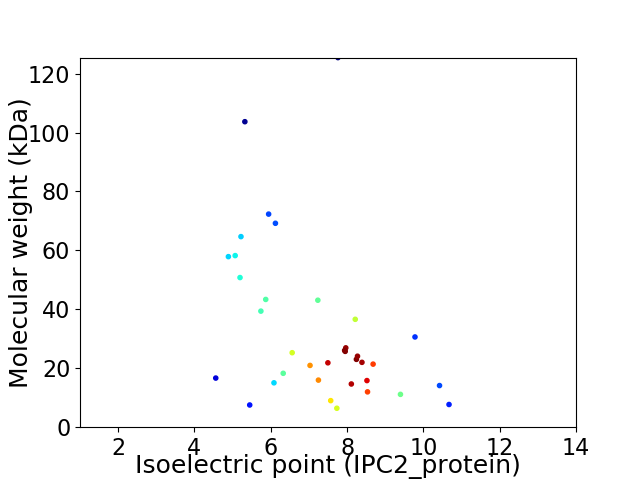
Virtual 2D-PAGE plot for 36 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q997H0|Q997H0_ADEB4 E4.2 OS=Bovine adenovirus 4 OX=70333 PE=4 SV=1
MM1 pKa = 7.0NQLSRR6 pKa = 11.84TFMGAAEE13 pKa = 4.2LVNYY17 pKa = 7.4FTDD20 pKa = 4.06LNSNQYY26 pKa = 11.07AIYY29 pKa = 10.25CVATSKK35 pKa = 11.09SLAYY39 pKa = 8.23WAKK42 pKa = 9.98GPFRR46 pKa = 11.84MGFRR50 pKa = 11.84FIGDD54 pKa = 3.46STVYY58 pKa = 10.96LLAQKK63 pKa = 10.73DD64 pKa = 3.97LFPEE68 pKa = 4.39FDD70 pKa = 4.16ALDD73 pKa = 3.72FLKK76 pKa = 10.23WANFSNMGMGFFAIEE91 pKa = 3.91WEE93 pKa = 4.11LLINSLVDD101 pKa = 3.67RR102 pKa = 11.84PMLTAEE108 pKa = 4.65VILIPGNTSHH118 pKa = 6.9EE119 pKa = 4.02SWEE122 pKa = 3.92FFKK125 pKa = 11.09YY126 pKa = 9.72YY127 pKa = 8.86WEE129 pKa = 4.33NYY131 pKa = 8.8FPYY134 pKa = 10.4FPIAPDD140 pKa = 3.33WEE142 pKa = 4.3
MM1 pKa = 7.0NQLSRR6 pKa = 11.84TFMGAAEE13 pKa = 4.2LVNYY17 pKa = 7.4FTDD20 pKa = 4.06LNSNQYY26 pKa = 11.07AIYY29 pKa = 10.25CVATSKK35 pKa = 11.09SLAYY39 pKa = 8.23WAKK42 pKa = 9.98GPFRR46 pKa = 11.84MGFRR50 pKa = 11.84FIGDD54 pKa = 3.46STVYY58 pKa = 10.96LLAQKK63 pKa = 10.73DD64 pKa = 3.97LFPEE68 pKa = 4.39FDD70 pKa = 4.16ALDD73 pKa = 3.72FLKK76 pKa = 10.23WANFSNMGMGFFAIEE91 pKa = 3.91WEE93 pKa = 4.11LLINSLVDD101 pKa = 3.67RR102 pKa = 11.84PMLTAEE108 pKa = 4.65VILIPGNTSHH118 pKa = 6.9EE119 pKa = 4.02SWEE122 pKa = 3.92FFKK125 pKa = 11.09YY126 pKa = 9.72YY127 pKa = 8.86WEE129 pKa = 4.33NYY131 pKa = 8.8FPYY134 pKa = 10.4FPIAPDD140 pKa = 3.33WEE142 pKa = 4.3
Molecular weight: 16.68 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q997I9|Q997I9_ADEB4 p32K OS=Bovine adenovirus 4 OX=70333 PE=4 SV=1
MM1 pKa = 7.27LTDD4 pKa = 4.43FYY6 pKa = 11.32NDD8 pKa = 3.48LMGGAYY14 pKa = 9.81RR15 pKa = 11.84RR16 pKa = 11.84TYY18 pKa = 10.69RR19 pKa = 11.84RR20 pKa = 11.84TRR22 pKa = 11.84KK23 pKa = 9.16AKK25 pKa = 9.52KK26 pKa = 9.67RR27 pKa = 11.84KK28 pKa = 9.06DD29 pKa = 3.3SRR31 pKa = 11.84RR32 pKa = 11.84SHH34 pKa = 6.62VNLKK38 pKa = 9.98KK39 pKa = 9.45RR40 pKa = 11.84TRR42 pKa = 11.84IARR45 pKa = 11.84NSNLYY50 pKa = 9.93SHH52 pKa = 7.08LRR54 pKa = 11.84LAPTAVNEE62 pKa = 4.16YY63 pKa = 9.23HH64 pKa = 6.53SKK66 pKa = 8.36PRR68 pKa = 11.84RR69 pKa = 11.84MSAVLHH75 pKa = 5.55MPISSVQLHH84 pKa = 5.63QIEE87 pKa = 4.58EE88 pKa = 4.38PKK90 pKa = 10.71HH91 pKa = 6.66SIASRR96 pKa = 11.84QFFNYY101 pKa = 9.06RR102 pKa = 11.84RR103 pKa = 11.84PPDD106 pKa = 3.67GEE108 pKa = 4.1EE109 pKa = 4.07DD110 pKa = 2.98VDD112 pKa = 3.89YY113 pKa = 10.85QAKK116 pKa = 10.3NKK118 pKa = 9.12WSLEE122 pKa = 3.96DD123 pKa = 3.52VISYY127 pKa = 8.68LQHH130 pKa = 6.12IPKK133 pKa = 9.77QVRR136 pKa = 11.84KK137 pKa = 10.33VILTSLFGATLGLIIDD153 pKa = 4.66ALLGGPWGLTTRR165 pKa = 11.84LLRR168 pKa = 11.84LIVSLVPGGKK178 pKa = 8.89ILLLALDD185 pKa = 3.77GLGYY189 pKa = 10.73FLGKK193 pKa = 10.2NNNPNLVAYY202 pKa = 10.13DD203 pKa = 3.57PDD205 pKa = 4.51LIKK208 pKa = 10.84FGTNIQRR215 pKa = 11.84NINGRR220 pKa = 11.84LTEE223 pKa = 4.93DD224 pKa = 2.81IVRR227 pKa = 11.84AAEE230 pKa = 3.94EE231 pKa = 3.87QLGGGFMRR239 pKa = 11.84TLAALLSAAASAGTHH254 pKa = 6.28LKK256 pKa = 10.0IALPAIPLAVIKK268 pKa = 9.89PFQRR272 pKa = 4.11
MM1 pKa = 7.27LTDD4 pKa = 4.43FYY6 pKa = 11.32NDD8 pKa = 3.48LMGGAYY14 pKa = 9.81RR15 pKa = 11.84RR16 pKa = 11.84TYY18 pKa = 10.69RR19 pKa = 11.84RR20 pKa = 11.84TRR22 pKa = 11.84KK23 pKa = 9.16AKK25 pKa = 9.52KK26 pKa = 9.67RR27 pKa = 11.84KK28 pKa = 9.06DD29 pKa = 3.3SRR31 pKa = 11.84RR32 pKa = 11.84SHH34 pKa = 6.62VNLKK38 pKa = 9.98KK39 pKa = 9.45RR40 pKa = 11.84TRR42 pKa = 11.84IARR45 pKa = 11.84NSNLYY50 pKa = 9.93SHH52 pKa = 7.08LRR54 pKa = 11.84LAPTAVNEE62 pKa = 4.16YY63 pKa = 9.23HH64 pKa = 6.53SKK66 pKa = 8.36PRR68 pKa = 11.84RR69 pKa = 11.84MSAVLHH75 pKa = 5.55MPISSVQLHH84 pKa = 5.63QIEE87 pKa = 4.58EE88 pKa = 4.38PKK90 pKa = 10.71HH91 pKa = 6.66SIASRR96 pKa = 11.84QFFNYY101 pKa = 9.06RR102 pKa = 11.84RR103 pKa = 11.84PPDD106 pKa = 3.67GEE108 pKa = 4.1EE109 pKa = 4.07DD110 pKa = 2.98VDD112 pKa = 3.89YY113 pKa = 10.85QAKK116 pKa = 10.3NKK118 pKa = 9.12WSLEE122 pKa = 3.96DD123 pKa = 3.52VISYY127 pKa = 8.68LQHH130 pKa = 6.12IPKK133 pKa = 9.77QVRR136 pKa = 11.84KK137 pKa = 10.33VILTSLFGATLGLIIDD153 pKa = 4.66ALLGGPWGLTTRR165 pKa = 11.84LLRR168 pKa = 11.84LIVSLVPGGKK178 pKa = 8.89ILLLALDD185 pKa = 3.77GLGYY189 pKa = 10.73FLGKK193 pKa = 10.2NNNPNLVAYY202 pKa = 10.13DD203 pKa = 3.57PDD205 pKa = 4.51LIKK208 pKa = 10.84FGTNIQRR215 pKa = 11.84NINGRR220 pKa = 11.84LTEE223 pKa = 4.93DD224 pKa = 2.81IVRR227 pKa = 11.84AAEE230 pKa = 3.94EE231 pKa = 3.87QLGGGFMRR239 pKa = 11.84TLAALLSAAASAGTHH254 pKa = 6.28LKK256 pKa = 10.0IALPAIPLAVIKK268 pKa = 9.89PFQRR272 pKa = 4.11
Molecular weight: 30.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
10427 |
54 |
1073 |
289.6 |
33.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.217 ± 0.292 | 2.58 ± 0.449 |
4.555 ± 0.216 | 5.63 ± 0.349 |
5.495 ± 0.226 | 4.555 ± 0.306 |
2.378 ± 0.214 | 5.965 ± 0.225 |
6.445 ± 0.613 | 10.674 ± 0.525 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.052 ± 0.171 | 6.272 ± 0.313 |
5.304 ± 0.259 | 4.066 ± 0.269 |
4.853 ± 0.386 | 7.413 ± 0.327 |
5.505 ± 0.329 | 5.265 ± 0.32 |
1.698 ± 0.194 | 4.076 ± 0.261 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
