
Algoriphagus sanaruensis
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Cyclobacteriaceae; Algoriphagus
Average proteome isoelectric point is 6.25
Get precalculated fractions of proteins
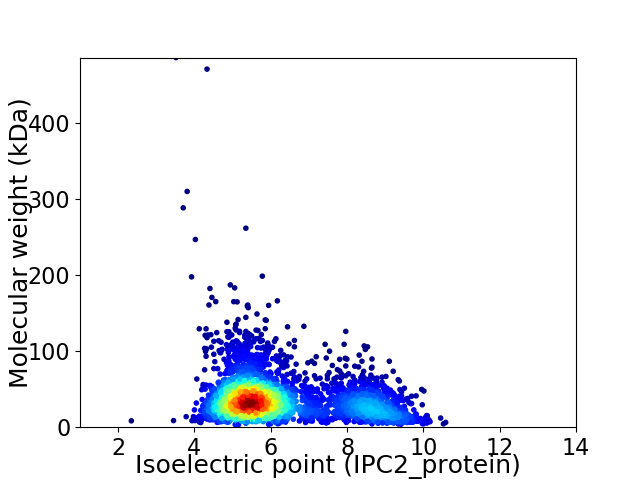
Virtual 2D-PAGE plot for 3377 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A142ES66|A0A142ES66_9BACT Glyco_trans_2-like domain-containing protein OS=Algoriphagus sanaruensis OX=1727163 GN=AO498_16075 PE=4 SV=1
MM1 pKa = 7.47KK2 pKa = 10.49SSIKK6 pKa = 10.25SIQYY10 pKa = 10.05RR11 pKa = 11.84LIFRR15 pKa = 11.84SILFCTWLGFSQFGYY30 pKa = 9.83SQGFNEE36 pKa = 4.43NEE38 pKa = 4.21WIFGSCGTGEE48 pKa = 3.84NSYY51 pKa = 11.46LSFGKK56 pKa = 10.57GGSANVQTLPGTVLIGKK73 pKa = 8.77NNNALAIDD81 pKa = 5.7PITGQPLFYY90 pKa = 10.35TNGVLVYY97 pKa = 9.62DD98 pKa = 4.87FSGSPIQGSAPGLNGDD114 pKa = 2.81IDD116 pKa = 4.06GRR118 pKa = 11.84QKK120 pKa = 10.8VATGFLEE127 pKa = 4.09YY128 pKa = 11.06NPDD131 pKa = 3.62GNKK134 pKa = 10.1LFYY137 pKa = 10.27IFYY140 pKa = 9.96LSPSGQLQYY149 pKa = 12.03ALVDD153 pKa = 3.7MNAPGQATGNEE164 pKa = 4.1RR165 pKa = 11.84PLGQVIEE172 pKa = 4.14KK173 pKa = 10.34DD174 pKa = 3.47QAIGNAEE181 pKa = 3.95GAVLVVKK188 pKa = 9.38TKK190 pKa = 10.7SSPSYY195 pKa = 10.42LISFSGGNLEE205 pKa = 3.9SRR207 pKa = 11.84RR208 pKa = 11.84FGSTAGSFSLTDD220 pKa = 3.35SEE222 pKa = 5.63GISFTPKK229 pKa = 10.55AIIFDD234 pKa = 3.91EE235 pKa = 4.66DD236 pKa = 3.21AGQLIVIPEE245 pKa = 4.05NPGEE249 pKa = 4.95DD250 pKa = 3.0ILVIPFDD257 pKa = 3.67TSTGNFGSPQAISEE271 pKa = 4.34SGGAEE276 pKa = 4.25DD277 pKa = 4.99IGGADD282 pKa = 4.69FSPDD286 pKa = 2.72GNFIYY291 pKa = 10.35FSRR294 pKa = 11.84GNQLFRR300 pKa = 11.84VPSSDD305 pKa = 4.45LNATPEE311 pKa = 4.36VIPTSPTNHH320 pKa = 6.26QIYY323 pKa = 10.03DD324 pKa = 3.8VKK326 pKa = 10.29TGPDD330 pKa = 3.2GQLYY334 pKa = 9.59YY335 pKa = 10.0IYY337 pKa = 9.73EE338 pKa = 4.21EE339 pKa = 4.54VVGGPQLIGRR349 pKa = 11.84VTNPNEE355 pKa = 3.66PDD357 pKa = 3.02IALVNLEE364 pKa = 4.35EE365 pKa = 5.21DD366 pKa = 4.12PFNGTDD372 pKa = 3.61FCGTIFPIFAPNTDD386 pKa = 3.38IDD388 pKa = 4.02PTVDD392 pKa = 4.1FTWDD396 pKa = 3.43PEE398 pKa = 4.37APCANNPVQLTSEE411 pKa = 4.23ITPEE415 pKa = 3.68NYY417 pKa = 9.81QPVSFNWEE425 pKa = 4.06FNPPLTDD432 pKa = 3.27SDD434 pKa = 4.64GNPLPADD441 pKa = 4.16FTQEE445 pKa = 3.95HH446 pKa = 6.46FLVPQDD452 pKa = 3.46ATEE455 pKa = 4.45GSSISVTLTVTFADD469 pKa = 3.85GTTRR473 pKa = 11.84NVTKK477 pKa = 10.37DD478 pKa = 2.94IPLSEE483 pKa = 4.65NNLEE487 pKa = 4.47ANFTPSDD494 pKa = 3.48TTLCEE499 pKa = 3.94SCIDD503 pKa = 5.08LDD505 pKa = 4.68PLLQAQSAAGQQDD518 pKa = 3.55GEE520 pKa = 4.38APGGGGQNYY529 pKa = 10.2EE530 pKa = 4.32YY531 pKa = 10.48FWSNKK536 pKa = 8.37RR537 pKa = 11.84DD538 pKa = 3.83EE539 pKa = 4.35GWLPKK544 pKa = 10.13GANEE548 pKa = 3.92VCRR551 pKa = 11.84PGLYY555 pKa = 8.81WVLVRR560 pKa = 11.84EE561 pKa = 4.76PGSSCYY567 pKa = 10.42AYY569 pKa = 10.62AEE571 pKa = 4.16IRR573 pKa = 11.84IKK575 pKa = 9.92MWDD578 pKa = 3.9IPDD581 pKa = 3.32QSNNIWYY588 pKa = 9.5FGDD591 pKa = 3.94GAGLDD596 pKa = 4.29FNPDD600 pKa = 3.6PDD602 pKa = 5.09DD603 pKa = 4.29EE604 pKa = 4.47NAPTPRR610 pKa = 11.84PIAQRR615 pKa = 11.84HH616 pKa = 5.04PQNIPAGTTTISDD629 pKa = 3.14QAGQVLFYY637 pKa = 10.81TDD639 pKa = 5.12GEE641 pKa = 4.94SVWDD645 pKa = 4.04LNGDD649 pKa = 4.35LMQNGDD655 pKa = 4.38NIGGDD660 pKa = 3.47NQATQGVIAVPVPTQPTLFYY680 pKa = 11.14LFTTQTNAGGTNTVSYY696 pKa = 10.75SLVDD700 pKa = 3.32IKK702 pKa = 11.21SEE704 pKa = 3.94NQTGVGNVVTKK715 pKa = 11.07DD716 pKa = 3.16NFLFSPATEE725 pKa = 3.81QSAALTAGDD734 pKa = 3.91TTWVLFHH741 pKa = 7.06EE742 pKa = 5.11LGNNTFRR749 pKa = 11.84SYY751 pKa = 10.99PVTVNGIGQPVLSSVGSNHH770 pKa = 6.07GFNSGVGSMKK780 pKa = 10.43FSPDD784 pKa = 2.97GTKK787 pKa = 10.28VAVTITEE794 pKa = 4.48GGCNKK799 pKa = 10.36LEE801 pKa = 3.9IFEE804 pKa = 4.84FDD806 pKa = 3.04QSTGEE811 pKa = 3.85LSEE814 pKa = 4.09YY815 pKa = 11.06ARR817 pKa = 11.84LDD819 pKa = 3.5LGCNGEE825 pKa = 4.28VYY827 pKa = 10.68GLEE830 pKa = 4.09FSQDD834 pKa = 2.96SDD836 pKa = 3.63RR837 pKa = 11.84VLVSYY842 pKa = 11.03RR843 pKa = 11.84NGGPGIEE850 pKa = 4.21EE851 pKa = 4.5FIIKK855 pKa = 9.77PVEE858 pKa = 4.15NNSTPNAPTCPSCFDD873 pKa = 3.65NASTRR878 pKa = 11.84AQRR881 pKa = 11.84EE882 pKa = 3.94ACILTTRR889 pKa = 11.84KK890 pKa = 9.82ALNGTSSLDD899 pKa = 3.53LGALQIASDD908 pKa = 3.87GQIYY912 pKa = 9.41VAVVGDD918 pKa = 3.68NRR920 pKa = 11.84IGQIQVGTGCTAASTFTQDD939 pKa = 4.28AVLPMPGTSNLGLPSFVQNSGSSIPEE965 pKa = 3.79PSLTIPTRR973 pKa = 11.84LCLDD977 pKa = 3.88PDD979 pKa = 4.05LGVSGLLEE987 pKa = 4.43GAGEE991 pKa = 4.18PDD993 pKa = 2.9IDD995 pKa = 4.19SYY997 pKa = 11.62FWTITHH1003 pKa = 7.02EE1004 pKa = 4.74DD1005 pKa = 3.46GTVIRR1010 pKa = 11.84NDD1012 pKa = 3.15FGGPGDD1018 pKa = 3.73QFQSLDD1024 pKa = 3.89QIFTLPGKK1032 pKa = 8.81YY1033 pKa = 7.88TVEE1036 pKa = 4.11LRR1038 pKa = 11.84VDD1040 pKa = 3.26RR1041 pKa = 11.84CGDD1044 pKa = 4.0PIYY1047 pKa = 11.15YY1048 pKa = 9.18EE1049 pKa = 3.91ATGEE1053 pKa = 4.31LEE1055 pKa = 4.62VVAPPTLTLAADD1067 pKa = 3.63ATLCAGNPVTLTAIDD1082 pKa = 4.97GYY1084 pKa = 11.44DD1085 pKa = 3.76PADD1088 pKa = 3.5GLYY1091 pKa = 10.73DD1092 pKa = 4.09FRR1094 pKa = 11.84WTNAAGQIIGDD1105 pKa = 4.1EE1106 pKa = 4.26NSNSITVDD1114 pKa = 2.96EE1115 pKa = 4.2EE1116 pKa = 3.98SIYY1119 pKa = 10.52RR1120 pKa = 11.84VEE1122 pKa = 3.9VSYY1125 pKa = 10.94RR1126 pKa = 11.84LPNGLSQEE1134 pKa = 4.14EE1135 pKa = 3.87ADD1137 pKa = 5.03LFEE1140 pKa = 4.91TCPSTAEE1147 pKa = 4.24VFVGPAFEE1155 pKa = 5.46FEE1157 pKa = 4.35LTQTATEE1164 pKa = 4.32VCFEE1168 pKa = 4.57EE1169 pKa = 5.2ISVTFAPDD1177 pKa = 3.52TPISGEE1183 pKa = 3.52WFYY1186 pKa = 11.63EE1187 pKa = 3.94RR1188 pKa = 11.84NGDD1191 pKa = 3.91GNRR1194 pKa = 11.84VSLGEE1199 pKa = 3.98FFEE1202 pKa = 5.15LEE1204 pKa = 4.0LWVNTLPGAGLYY1216 pKa = 10.24DD1217 pKa = 3.45IYY1219 pKa = 10.92FVTEE1223 pKa = 3.97DD1224 pKa = 5.06PILAGCTVEE1233 pKa = 6.39KK1234 pKa = 10.61RR1235 pKa = 11.84LQLQVNEE1242 pKa = 4.73LPLFVPAQTSPATDD1256 pKa = 3.52CATADD1261 pKa = 3.44GGFEE1265 pKa = 3.58ITMQADD1271 pKa = 3.25AAEE1274 pKa = 4.11VRR1276 pKa = 11.84IPEE1279 pKa = 4.35LSLSFTNVSAGDD1291 pKa = 3.97VIPVANVLPGVYY1303 pKa = 8.75TVEE1306 pKa = 4.44AEE1308 pKa = 4.11SSAGCFYY1315 pKa = 10.82SGSVTVEE1322 pKa = 3.74NANPPSGLAFTVATTDD1338 pKa = 3.43EE1339 pKa = 4.72TCASTGVNPGIISITFTSGTAQTGTYY1365 pKa = 9.35TITRR1369 pKa = 11.84QGDD1372 pKa = 3.69GSVFTGPLPNQAAFDD1387 pKa = 3.95VEE1389 pKa = 4.63VPYY1392 pKa = 10.9GSYY1395 pKa = 10.39LVEE1398 pKa = 3.96VADD1401 pKa = 5.08ASGCSVPDD1409 pKa = 3.78PSLYY1413 pKa = 10.77SVAQKK1418 pKa = 10.75FEE1420 pKa = 4.54VVFSVPTSVEE1430 pKa = 3.51ACEE1433 pKa = 4.58IFAFTPIGPNTLNYY1447 pKa = 10.6LVTNSSGTVINPDD1460 pKa = 3.2ANGEE1464 pKa = 4.09YY1465 pKa = 10.72LLTSSGIYY1473 pKa = 7.62TVRR1476 pKa = 11.84GEE1478 pKa = 4.42DD1479 pKa = 3.9PNGVDD1484 pKa = 4.62CPRR1487 pKa = 11.84EE1488 pKa = 3.68IQMDD1492 pKa = 3.8VTLSQPIDD1500 pKa = 3.85FSVSGPIIDD1509 pKa = 4.11CQAGVRR1515 pKa = 11.84FEE1517 pKa = 5.06AVLNNALPQDD1527 pKa = 4.74VIFLWKK1533 pKa = 10.14DD1534 pKa = 3.22DD1535 pKa = 3.96LGVIVGRR1542 pKa = 11.84RR1543 pKa = 11.84QTFTPSRR1550 pKa = 11.84DD1551 pKa = 3.09GTYY1554 pKa = 9.97TLEE1557 pKa = 4.08VQPASGGLCTTPAQSFTAEE1576 pKa = 4.09ILDD1579 pKa = 3.87EE1580 pKa = 4.46AVTVALEE1587 pKa = 4.02ADD1589 pKa = 4.79PYY1591 pKa = 10.9CISQTSTVIDD1601 pKa = 3.68ILADD1605 pKa = 3.67LQNVAEE1611 pKa = 4.51IEE1613 pKa = 4.08WFAVSGGTRR1622 pKa = 11.84TRR1624 pKa = 11.84LPNFDD1629 pKa = 4.1NLSSITVTDD1638 pKa = 3.32EE1639 pKa = 3.81GTYY1642 pKa = 10.16EE1643 pKa = 3.95ALLRR1647 pKa = 11.84SAQGCEE1653 pKa = 3.77LGRR1656 pKa = 11.84ANIVVVKK1663 pKa = 8.74STIIPPVVPTTPITICAVEE1682 pKa = 5.17GITTSINPGNYY1693 pKa = 9.87DD1694 pKa = 3.9FYY1696 pKa = 11.16SWRR1699 pKa = 11.84LNGVEE1704 pKa = 4.27ISQDD1708 pKa = 3.51AIFSPTLPGEE1718 pKa = 4.13YY1719 pKa = 9.91EE1720 pKa = 4.11LRR1722 pKa = 11.84VSDD1725 pKa = 4.64NLGCEE1730 pKa = 4.0YY1731 pKa = 10.76VANFTILEE1739 pKa = 4.16DD1740 pKa = 3.32CRR1742 pKa = 11.84LRR1744 pKa = 11.84VSYY1747 pKa = 10.72PNGVVLGDD1755 pKa = 3.66PNRR1758 pKa = 11.84NFILYY1763 pKa = 9.88ANEE1766 pKa = 4.75FIDD1769 pKa = 3.89EE1770 pKa = 4.39VEE1772 pKa = 4.03VFIFNRR1778 pKa = 11.84WGEE1781 pKa = 4.09LIFYY1785 pKa = 9.95CEE1787 pKa = 4.46HH1788 pKa = 6.84EE1789 pKa = 4.33NLEE1792 pKa = 4.72PKK1794 pKa = 9.44TAFCPWDD1801 pKa = 3.46GQVNGKK1807 pKa = 8.85FVPNGTYY1814 pKa = 9.86AVKK1817 pKa = 10.85VKK1819 pKa = 9.1FTSRR1823 pKa = 11.84NQNLTQTEE1831 pKa = 4.52TKK1833 pKa = 10.3AITIIQQ1839 pKa = 3.58
MM1 pKa = 7.47KK2 pKa = 10.49SSIKK6 pKa = 10.25SIQYY10 pKa = 10.05RR11 pKa = 11.84LIFRR15 pKa = 11.84SILFCTWLGFSQFGYY30 pKa = 9.83SQGFNEE36 pKa = 4.43NEE38 pKa = 4.21WIFGSCGTGEE48 pKa = 3.84NSYY51 pKa = 11.46LSFGKK56 pKa = 10.57GGSANVQTLPGTVLIGKK73 pKa = 8.77NNNALAIDD81 pKa = 5.7PITGQPLFYY90 pKa = 10.35TNGVLVYY97 pKa = 9.62DD98 pKa = 4.87FSGSPIQGSAPGLNGDD114 pKa = 2.81IDD116 pKa = 4.06GRR118 pKa = 11.84QKK120 pKa = 10.8VATGFLEE127 pKa = 4.09YY128 pKa = 11.06NPDD131 pKa = 3.62GNKK134 pKa = 10.1LFYY137 pKa = 10.27IFYY140 pKa = 9.96LSPSGQLQYY149 pKa = 12.03ALVDD153 pKa = 3.7MNAPGQATGNEE164 pKa = 4.1RR165 pKa = 11.84PLGQVIEE172 pKa = 4.14KK173 pKa = 10.34DD174 pKa = 3.47QAIGNAEE181 pKa = 3.95GAVLVVKK188 pKa = 9.38TKK190 pKa = 10.7SSPSYY195 pKa = 10.42LISFSGGNLEE205 pKa = 3.9SRR207 pKa = 11.84RR208 pKa = 11.84FGSTAGSFSLTDD220 pKa = 3.35SEE222 pKa = 5.63GISFTPKK229 pKa = 10.55AIIFDD234 pKa = 3.91EE235 pKa = 4.66DD236 pKa = 3.21AGQLIVIPEE245 pKa = 4.05NPGEE249 pKa = 4.95DD250 pKa = 3.0ILVIPFDD257 pKa = 3.67TSTGNFGSPQAISEE271 pKa = 4.34SGGAEE276 pKa = 4.25DD277 pKa = 4.99IGGADD282 pKa = 4.69FSPDD286 pKa = 2.72GNFIYY291 pKa = 10.35FSRR294 pKa = 11.84GNQLFRR300 pKa = 11.84VPSSDD305 pKa = 4.45LNATPEE311 pKa = 4.36VIPTSPTNHH320 pKa = 6.26QIYY323 pKa = 10.03DD324 pKa = 3.8VKK326 pKa = 10.29TGPDD330 pKa = 3.2GQLYY334 pKa = 9.59YY335 pKa = 10.0IYY337 pKa = 9.73EE338 pKa = 4.21EE339 pKa = 4.54VVGGPQLIGRR349 pKa = 11.84VTNPNEE355 pKa = 3.66PDD357 pKa = 3.02IALVNLEE364 pKa = 4.35EE365 pKa = 5.21DD366 pKa = 4.12PFNGTDD372 pKa = 3.61FCGTIFPIFAPNTDD386 pKa = 3.38IDD388 pKa = 4.02PTVDD392 pKa = 4.1FTWDD396 pKa = 3.43PEE398 pKa = 4.37APCANNPVQLTSEE411 pKa = 4.23ITPEE415 pKa = 3.68NYY417 pKa = 9.81QPVSFNWEE425 pKa = 4.06FNPPLTDD432 pKa = 3.27SDD434 pKa = 4.64GNPLPADD441 pKa = 4.16FTQEE445 pKa = 3.95HH446 pKa = 6.46FLVPQDD452 pKa = 3.46ATEE455 pKa = 4.45GSSISVTLTVTFADD469 pKa = 3.85GTTRR473 pKa = 11.84NVTKK477 pKa = 10.37DD478 pKa = 2.94IPLSEE483 pKa = 4.65NNLEE487 pKa = 4.47ANFTPSDD494 pKa = 3.48TTLCEE499 pKa = 3.94SCIDD503 pKa = 5.08LDD505 pKa = 4.68PLLQAQSAAGQQDD518 pKa = 3.55GEE520 pKa = 4.38APGGGGQNYY529 pKa = 10.2EE530 pKa = 4.32YY531 pKa = 10.48FWSNKK536 pKa = 8.37RR537 pKa = 11.84DD538 pKa = 3.83EE539 pKa = 4.35GWLPKK544 pKa = 10.13GANEE548 pKa = 3.92VCRR551 pKa = 11.84PGLYY555 pKa = 8.81WVLVRR560 pKa = 11.84EE561 pKa = 4.76PGSSCYY567 pKa = 10.42AYY569 pKa = 10.62AEE571 pKa = 4.16IRR573 pKa = 11.84IKK575 pKa = 9.92MWDD578 pKa = 3.9IPDD581 pKa = 3.32QSNNIWYY588 pKa = 9.5FGDD591 pKa = 3.94GAGLDD596 pKa = 4.29FNPDD600 pKa = 3.6PDD602 pKa = 5.09DD603 pKa = 4.29EE604 pKa = 4.47NAPTPRR610 pKa = 11.84PIAQRR615 pKa = 11.84HH616 pKa = 5.04PQNIPAGTTTISDD629 pKa = 3.14QAGQVLFYY637 pKa = 10.81TDD639 pKa = 5.12GEE641 pKa = 4.94SVWDD645 pKa = 4.04LNGDD649 pKa = 4.35LMQNGDD655 pKa = 4.38NIGGDD660 pKa = 3.47NQATQGVIAVPVPTQPTLFYY680 pKa = 11.14LFTTQTNAGGTNTVSYY696 pKa = 10.75SLVDD700 pKa = 3.32IKK702 pKa = 11.21SEE704 pKa = 3.94NQTGVGNVVTKK715 pKa = 11.07DD716 pKa = 3.16NFLFSPATEE725 pKa = 3.81QSAALTAGDD734 pKa = 3.91TTWVLFHH741 pKa = 7.06EE742 pKa = 5.11LGNNTFRR749 pKa = 11.84SYY751 pKa = 10.99PVTVNGIGQPVLSSVGSNHH770 pKa = 6.07GFNSGVGSMKK780 pKa = 10.43FSPDD784 pKa = 2.97GTKK787 pKa = 10.28VAVTITEE794 pKa = 4.48GGCNKK799 pKa = 10.36LEE801 pKa = 3.9IFEE804 pKa = 4.84FDD806 pKa = 3.04QSTGEE811 pKa = 3.85LSEE814 pKa = 4.09YY815 pKa = 11.06ARR817 pKa = 11.84LDD819 pKa = 3.5LGCNGEE825 pKa = 4.28VYY827 pKa = 10.68GLEE830 pKa = 4.09FSQDD834 pKa = 2.96SDD836 pKa = 3.63RR837 pKa = 11.84VLVSYY842 pKa = 11.03RR843 pKa = 11.84NGGPGIEE850 pKa = 4.21EE851 pKa = 4.5FIIKK855 pKa = 9.77PVEE858 pKa = 4.15NNSTPNAPTCPSCFDD873 pKa = 3.65NASTRR878 pKa = 11.84AQRR881 pKa = 11.84EE882 pKa = 3.94ACILTTRR889 pKa = 11.84KK890 pKa = 9.82ALNGTSSLDD899 pKa = 3.53LGALQIASDD908 pKa = 3.87GQIYY912 pKa = 9.41VAVVGDD918 pKa = 3.68NRR920 pKa = 11.84IGQIQVGTGCTAASTFTQDD939 pKa = 4.28AVLPMPGTSNLGLPSFVQNSGSSIPEE965 pKa = 3.79PSLTIPTRR973 pKa = 11.84LCLDD977 pKa = 3.88PDD979 pKa = 4.05LGVSGLLEE987 pKa = 4.43GAGEE991 pKa = 4.18PDD993 pKa = 2.9IDD995 pKa = 4.19SYY997 pKa = 11.62FWTITHH1003 pKa = 7.02EE1004 pKa = 4.74DD1005 pKa = 3.46GTVIRR1010 pKa = 11.84NDD1012 pKa = 3.15FGGPGDD1018 pKa = 3.73QFQSLDD1024 pKa = 3.89QIFTLPGKK1032 pKa = 8.81YY1033 pKa = 7.88TVEE1036 pKa = 4.11LRR1038 pKa = 11.84VDD1040 pKa = 3.26RR1041 pKa = 11.84CGDD1044 pKa = 4.0PIYY1047 pKa = 11.15YY1048 pKa = 9.18EE1049 pKa = 3.91ATGEE1053 pKa = 4.31LEE1055 pKa = 4.62VVAPPTLTLAADD1067 pKa = 3.63ATLCAGNPVTLTAIDD1082 pKa = 4.97GYY1084 pKa = 11.44DD1085 pKa = 3.76PADD1088 pKa = 3.5GLYY1091 pKa = 10.73DD1092 pKa = 4.09FRR1094 pKa = 11.84WTNAAGQIIGDD1105 pKa = 4.1EE1106 pKa = 4.26NSNSITVDD1114 pKa = 2.96EE1115 pKa = 4.2EE1116 pKa = 3.98SIYY1119 pKa = 10.52RR1120 pKa = 11.84VEE1122 pKa = 3.9VSYY1125 pKa = 10.94RR1126 pKa = 11.84LPNGLSQEE1134 pKa = 4.14EE1135 pKa = 3.87ADD1137 pKa = 5.03LFEE1140 pKa = 4.91TCPSTAEE1147 pKa = 4.24VFVGPAFEE1155 pKa = 5.46FEE1157 pKa = 4.35LTQTATEE1164 pKa = 4.32VCFEE1168 pKa = 4.57EE1169 pKa = 5.2ISVTFAPDD1177 pKa = 3.52TPISGEE1183 pKa = 3.52WFYY1186 pKa = 11.63EE1187 pKa = 3.94RR1188 pKa = 11.84NGDD1191 pKa = 3.91GNRR1194 pKa = 11.84VSLGEE1199 pKa = 3.98FFEE1202 pKa = 5.15LEE1204 pKa = 4.0LWVNTLPGAGLYY1216 pKa = 10.24DD1217 pKa = 3.45IYY1219 pKa = 10.92FVTEE1223 pKa = 3.97DD1224 pKa = 5.06PILAGCTVEE1233 pKa = 6.39KK1234 pKa = 10.61RR1235 pKa = 11.84LQLQVNEE1242 pKa = 4.73LPLFVPAQTSPATDD1256 pKa = 3.52CATADD1261 pKa = 3.44GGFEE1265 pKa = 3.58ITMQADD1271 pKa = 3.25AAEE1274 pKa = 4.11VRR1276 pKa = 11.84IPEE1279 pKa = 4.35LSLSFTNVSAGDD1291 pKa = 3.97VIPVANVLPGVYY1303 pKa = 8.75TVEE1306 pKa = 4.44AEE1308 pKa = 4.11SSAGCFYY1315 pKa = 10.82SGSVTVEE1322 pKa = 3.74NANPPSGLAFTVATTDD1338 pKa = 3.43EE1339 pKa = 4.72TCASTGVNPGIISITFTSGTAQTGTYY1365 pKa = 9.35TITRR1369 pKa = 11.84QGDD1372 pKa = 3.69GSVFTGPLPNQAAFDD1387 pKa = 3.95VEE1389 pKa = 4.63VPYY1392 pKa = 10.9GSYY1395 pKa = 10.39LVEE1398 pKa = 3.96VADD1401 pKa = 5.08ASGCSVPDD1409 pKa = 3.78PSLYY1413 pKa = 10.77SVAQKK1418 pKa = 10.75FEE1420 pKa = 4.54VVFSVPTSVEE1430 pKa = 3.51ACEE1433 pKa = 4.58IFAFTPIGPNTLNYY1447 pKa = 10.6LVTNSSGTVINPDD1460 pKa = 3.2ANGEE1464 pKa = 4.09YY1465 pKa = 10.72LLTSSGIYY1473 pKa = 7.62TVRR1476 pKa = 11.84GEE1478 pKa = 4.42DD1479 pKa = 3.9PNGVDD1484 pKa = 4.62CPRR1487 pKa = 11.84EE1488 pKa = 3.68IQMDD1492 pKa = 3.8VTLSQPIDD1500 pKa = 3.85FSVSGPIIDD1509 pKa = 4.11CQAGVRR1515 pKa = 11.84FEE1517 pKa = 5.06AVLNNALPQDD1527 pKa = 4.74VIFLWKK1533 pKa = 10.14DD1534 pKa = 3.22DD1535 pKa = 3.96LGVIVGRR1542 pKa = 11.84RR1543 pKa = 11.84QTFTPSRR1550 pKa = 11.84DD1551 pKa = 3.09GTYY1554 pKa = 9.97TLEE1557 pKa = 4.08VQPASGGLCTTPAQSFTAEE1576 pKa = 4.09ILDD1579 pKa = 3.87EE1580 pKa = 4.46AVTVALEE1587 pKa = 4.02ADD1589 pKa = 4.79PYY1591 pKa = 10.9CISQTSTVIDD1601 pKa = 3.68ILADD1605 pKa = 3.67LQNVAEE1611 pKa = 4.51IEE1613 pKa = 4.08WFAVSGGTRR1622 pKa = 11.84TRR1624 pKa = 11.84LPNFDD1629 pKa = 4.1NLSSITVTDD1638 pKa = 3.32EE1639 pKa = 3.81GTYY1642 pKa = 10.16EE1643 pKa = 3.95ALLRR1647 pKa = 11.84SAQGCEE1653 pKa = 3.77LGRR1656 pKa = 11.84ANIVVVKK1663 pKa = 8.74STIIPPVVPTTPITICAVEE1682 pKa = 5.17GITTSINPGNYY1693 pKa = 9.87DD1694 pKa = 3.9FYY1696 pKa = 11.16SWRR1699 pKa = 11.84LNGVEE1704 pKa = 4.27ISQDD1708 pKa = 3.51AIFSPTLPGEE1718 pKa = 4.13YY1719 pKa = 9.91EE1720 pKa = 4.11LRR1722 pKa = 11.84VSDD1725 pKa = 4.64NLGCEE1730 pKa = 4.0YY1731 pKa = 10.76VANFTILEE1739 pKa = 4.16DD1740 pKa = 3.32CRR1742 pKa = 11.84LRR1744 pKa = 11.84VSYY1747 pKa = 10.72PNGVVLGDD1755 pKa = 3.66PNRR1758 pKa = 11.84NFILYY1763 pKa = 9.88ANEE1766 pKa = 4.75FIDD1769 pKa = 3.89EE1770 pKa = 4.39VEE1772 pKa = 4.03VFIFNRR1778 pKa = 11.84WGEE1781 pKa = 4.09LIFYY1785 pKa = 9.95CEE1787 pKa = 4.46HH1788 pKa = 6.84EE1789 pKa = 4.33NLEE1792 pKa = 4.72PKK1794 pKa = 9.44TAFCPWDD1801 pKa = 3.46GQVNGKK1807 pKa = 8.85FVPNGTYY1814 pKa = 9.86AVKK1817 pKa = 10.85VKK1819 pKa = 9.1FTSRR1823 pKa = 11.84NQNLTQTEE1831 pKa = 4.52TKK1833 pKa = 10.3AITIIQQ1839 pKa = 3.58
Molecular weight: 197.79 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A142EKI2|A0A142EKI2_9BACT Peptidase M23 OS=Algoriphagus sanaruensis OX=1727163 GN=AO498_04410 PE=4 SV=1
MM1 pKa = 6.99SQEE4 pKa = 3.91EE5 pKa = 4.29VLGQLRR11 pKa = 11.84AVTRR15 pKa = 11.84EE16 pKa = 3.72ANFLDD21 pKa = 4.96ARR23 pKa = 11.84TQTLADD29 pKa = 3.45VKK31 pKa = 11.13FNGLVGRR38 pKa = 11.84QKK40 pKa = 10.52FRR42 pKa = 11.84ISRR45 pKa = 11.84VIRR48 pKa = 11.84RR49 pKa = 11.84GEE51 pKa = 3.96TFLPLIEE58 pKa = 4.47GRR60 pKa = 11.84VEE62 pKa = 3.65KK63 pKa = 10.01TPRR66 pKa = 11.84GSIIFLAYY74 pKa = 10.4RR75 pKa = 11.84LFPVALFFLIFWSVVLVSFAIFYY98 pKa = 11.11GLALEE103 pKa = 4.37NWQNAMICSLLAVVNYY119 pKa = 6.71MLCVFFFHH127 pKa = 6.65RR128 pKa = 11.84QVKK131 pKa = 9.15ASRR134 pKa = 11.84AIFHH138 pKa = 5.81QLINFHH144 pKa = 6.84LKK146 pKa = 10.76GKK148 pKa = 10.0DD149 pKa = 3.02
MM1 pKa = 6.99SQEE4 pKa = 3.91EE5 pKa = 4.29VLGQLRR11 pKa = 11.84AVTRR15 pKa = 11.84EE16 pKa = 3.72ANFLDD21 pKa = 4.96ARR23 pKa = 11.84TQTLADD29 pKa = 3.45VKK31 pKa = 11.13FNGLVGRR38 pKa = 11.84QKK40 pKa = 10.52FRR42 pKa = 11.84ISRR45 pKa = 11.84VIRR48 pKa = 11.84RR49 pKa = 11.84GEE51 pKa = 3.96TFLPLIEE58 pKa = 4.47GRR60 pKa = 11.84VEE62 pKa = 3.65KK63 pKa = 10.01TPRR66 pKa = 11.84GSIIFLAYY74 pKa = 10.4RR75 pKa = 11.84LFPVALFFLIFWSVVLVSFAIFYY98 pKa = 11.11GLALEE103 pKa = 4.37NWQNAMICSLLAVVNYY119 pKa = 6.71MLCVFFFHH127 pKa = 6.65RR128 pKa = 11.84QVKK131 pKa = 9.15ASRR134 pKa = 11.84AIFHH138 pKa = 5.81QLINFHH144 pKa = 6.84LKK146 pKa = 10.76GKK148 pKa = 10.0DD149 pKa = 3.02
Molecular weight: 17.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1184144 |
29 |
4663 |
350.6 |
39.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.821 ± 0.052 | 0.656 ± 0.015 |
5.149 ± 0.03 | 6.904 ± 0.042 |
5.378 ± 0.035 | 7.355 ± 0.062 |
1.85 ± 0.026 | 7.106 ± 0.035 |
6.506 ± 0.045 | 10.032 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.309 ± 0.023 | 4.725 ± 0.033 |
4.148 ± 0.024 | 4.042 ± 0.03 |
4.075 ± 0.031 | 6.742 ± 0.031 |
5.159 ± 0.068 | 6.179 ± 0.033 |
1.35 ± 0.017 | 3.514 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
