
Candidatus Tremblaya phenacola
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Betaproteobacteria incertae sedis; Candidatus Tremblaya
Average proteome isoelectric point is 8.52
Get precalculated fractions of proteins
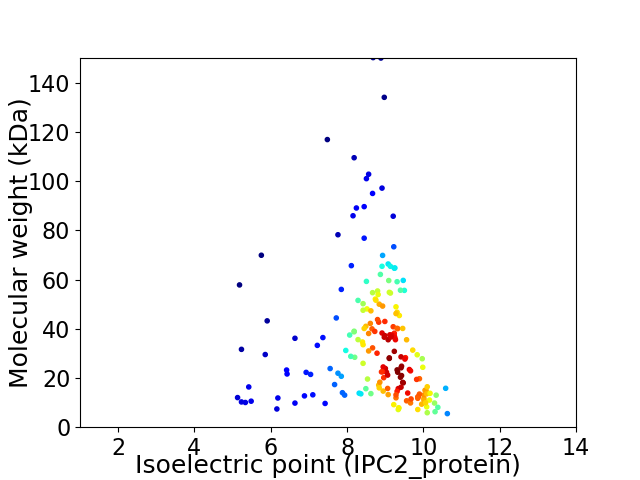
Virtual 2D-PAGE plot for 194 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2G0V6U6|A0A2G0V6U6_9PROT 30S ribosomal protein S11 OS=Candidatus Tremblaya phenacola OX=1010676 GN=rpsK PE=3 SV=1
MM1 pKa = 7.37YY2 pKa = 8.12EE3 pKa = 3.89ANEE6 pKa = 4.16ANEE9 pKa = 4.16TMNRR13 pKa = 11.84LTAEE17 pKa = 4.14HH18 pKa = 5.84TTAFHH23 pKa = 6.99PNLLRR28 pKa = 11.84DD29 pKa = 4.06TFPTEE34 pKa = 3.84SSHH37 pKa = 6.78TYY39 pKa = 10.56SLTNILFCLLLTLTPLPTIQNHH61 pKa = 5.87LLIIPEE67 pKa = 4.27INPEE71 pKa = 3.94HH72 pKa = 7.15PYY74 pKa = 10.63VLPRR78 pKa = 11.84YY79 pKa = 8.47CLRR82 pKa = 11.84VMMSNKK88 pKa = 9.7CYY90 pKa = 10.51CLIDD94 pKa = 3.53IDD96 pKa = 5.17SPLEE100 pKa = 3.82PEE102 pKa = 4.2RR103 pKa = 11.84AA104 pKa = 3.49
MM1 pKa = 7.37YY2 pKa = 8.12EE3 pKa = 3.89ANEE6 pKa = 4.16ANEE9 pKa = 4.16TMNRR13 pKa = 11.84LTAEE17 pKa = 4.14HH18 pKa = 5.84TTAFHH23 pKa = 6.99PNLLRR28 pKa = 11.84DD29 pKa = 4.06TFPTEE34 pKa = 3.84SSHH37 pKa = 6.78TYY39 pKa = 10.56SLTNILFCLLLTLTPLPTIQNHH61 pKa = 5.87LLIIPEE67 pKa = 4.27INPEE71 pKa = 3.94HH72 pKa = 7.15PYY74 pKa = 10.63VLPRR78 pKa = 11.84YY79 pKa = 8.47CLRR82 pKa = 11.84VMMSNKK88 pKa = 9.7CYY90 pKa = 10.51CLIDD94 pKa = 3.53IDD96 pKa = 5.17SPLEE100 pKa = 3.82PEE102 pKa = 4.2RR103 pKa = 11.84AA104 pKa = 3.49
Molecular weight: 12.03 kDa
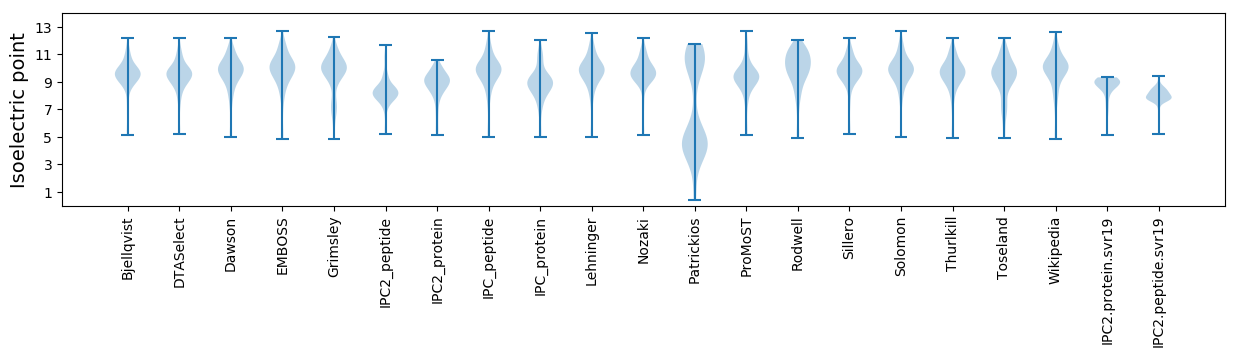
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2G0V6X8|A0A2G0V6X8_9PROT IGP synthase cyclase subunit OS=Candidatus Tremblaya phenacola OX=1010676 GN=hisF PE=3 SV=1
MM1 pKa = 7.71LIKK4 pKa = 10.66NSPYY8 pKa = 10.43PMLLRR13 pKa = 11.84TEE15 pKa = 4.45RR16 pKa = 11.84KK17 pKa = 9.24SSGRR21 pKa = 11.84NNIGTITVRR30 pKa = 11.84HH31 pKa = 6.33RR32 pKa = 11.84GCGHH36 pKa = 5.83KK37 pKa = 10.09RR38 pKa = 11.84LYY40 pKa = 9.5RR41 pKa = 11.84TIDD44 pKa = 3.77FYY46 pKa = 11.68RR47 pKa = 11.84NKK49 pKa = 10.65DD50 pKa = 3.65YY51 pKa = 11.18ILGKK55 pKa = 8.89VEE57 pKa = 4.02KK58 pKa = 10.72NEE60 pKa = 3.79YY61 pKa = 10.38DD62 pKa = 3.56PNRR65 pKa = 11.84SANIVLVLYY74 pKa = 10.83ADD76 pKa = 3.82GEE78 pKa = 4.19RR79 pKa = 11.84RR80 pKa = 11.84YY81 pKa = 10.23IIAPNRR87 pKa = 11.84LPIGSVIVSGRR98 pKa = 11.84SVPSQIGNSLSVGLIYY114 pKa = 10.77SGTNVHH120 pKa = 6.82CIEE123 pKa = 4.55LVPANGAKK131 pKa = 10.56LSRR134 pKa = 11.84AAGSYY139 pKa = 8.86ATIISNASHH148 pKa = 5.29YY149 pKa = 8.16TQVRR153 pKa = 11.84LMSGEE158 pKa = 3.77VRR160 pKa = 11.84LINKK164 pKa = 7.52EE165 pKa = 3.67CRR167 pKa = 11.84ATVGIVGNIKK177 pKa = 10.41HH178 pKa = 6.01SLSRR182 pKa = 11.84LDD184 pKa = 3.41KK185 pKa = 10.01AGKK188 pKa = 9.76RR189 pKa = 11.84RR190 pKa = 11.84WLGIRR195 pKa = 11.84PTVRR199 pKa = 11.84GVAMNPIDD207 pKa = 4.06HH208 pKa = 6.89PHH210 pKa = 6.59GGGEE214 pKa = 4.38GKK216 pKa = 6.94TTSGRR221 pKa = 11.84HH222 pKa = 5.12PVSPWGTPTKK232 pKa = 10.67GFKK235 pKa = 8.99TRR237 pKa = 11.84NNKK240 pKa = 9.51QSDD243 pKa = 3.96SYY245 pKa = 11.06IINKK249 pKa = 9.02RR250 pKa = 11.84RR251 pKa = 3.17
MM1 pKa = 7.71LIKK4 pKa = 10.66NSPYY8 pKa = 10.43PMLLRR13 pKa = 11.84TEE15 pKa = 4.45RR16 pKa = 11.84KK17 pKa = 9.24SSGRR21 pKa = 11.84NNIGTITVRR30 pKa = 11.84HH31 pKa = 6.33RR32 pKa = 11.84GCGHH36 pKa = 5.83KK37 pKa = 10.09RR38 pKa = 11.84LYY40 pKa = 9.5RR41 pKa = 11.84TIDD44 pKa = 3.77FYY46 pKa = 11.68RR47 pKa = 11.84NKK49 pKa = 10.65DD50 pKa = 3.65YY51 pKa = 11.18ILGKK55 pKa = 8.89VEE57 pKa = 4.02KK58 pKa = 10.72NEE60 pKa = 3.79YY61 pKa = 10.38DD62 pKa = 3.56PNRR65 pKa = 11.84SANIVLVLYY74 pKa = 10.83ADD76 pKa = 3.82GEE78 pKa = 4.19RR79 pKa = 11.84RR80 pKa = 11.84YY81 pKa = 10.23IIAPNRR87 pKa = 11.84LPIGSVIVSGRR98 pKa = 11.84SVPSQIGNSLSVGLIYY114 pKa = 10.77SGTNVHH120 pKa = 6.82CIEE123 pKa = 4.55LVPANGAKK131 pKa = 10.56LSRR134 pKa = 11.84AAGSYY139 pKa = 8.86ATIISNASHH148 pKa = 5.29YY149 pKa = 8.16TQVRR153 pKa = 11.84LMSGEE158 pKa = 3.77VRR160 pKa = 11.84LINKK164 pKa = 7.52EE165 pKa = 3.67CRR167 pKa = 11.84ATVGIVGNIKK177 pKa = 10.41HH178 pKa = 6.01SLSRR182 pKa = 11.84LDD184 pKa = 3.41KK185 pKa = 10.01AGKK188 pKa = 9.76RR189 pKa = 11.84RR190 pKa = 11.84WLGIRR195 pKa = 11.84PTVRR199 pKa = 11.84GVAMNPIDD207 pKa = 4.06HH208 pKa = 6.89PHH210 pKa = 6.59GGGEE214 pKa = 4.38GKK216 pKa = 6.94TTSGRR221 pKa = 11.84HH222 pKa = 5.12PVSPWGTPTKK232 pKa = 10.67GFKK235 pKa = 8.99TRR237 pKa = 11.84NNKK240 pKa = 9.51QSDD243 pKa = 3.96SYY245 pKa = 11.06IINKK249 pKa = 9.02RR250 pKa = 11.84RR251 pKa = 3.17
Molecular weight: 27.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
60884 |
46 |
1351 |
313.8 |
35.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.139 ± 0.131 | 1.703 ± 0.06 |
3.64 ± 0.087 | 4.74 ± 0.136 |
3.446 ± 0.094 | 5.775 ± 0.118 |
2.247 ± 0.074 | 9.668 ± 0.214 |
8.033 ± 0.141 | 10.592 ± 0.165 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.088 ± 0.057 | 5.921 ± 0.116 |
3.232 ± 0.076 | 2.505 ± 0.086 |
5.092 ± 0.101 | 8.546 ± 0.137 |
5.709 ± 0.11 | 6.747 ± 0.167 |
0.828 ± 0.062 | 4.349 ± 0.141 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
