
Encephalitozoon cuniculi (strain GB-M1) (Microsporidian parasite)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Fungi incertae sedis; Microsporidia; Apansporoblastina; Unikaryonidae; Encephalitozoon; Encephalitozoon cuniculi
Average proteome isoelectric point is 6.83
Get precalculated fractions of proteins
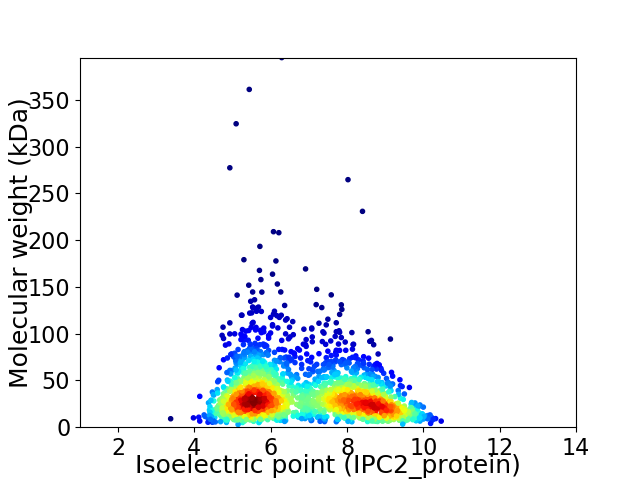
Virtual 2D-PAGE plot for 2041 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
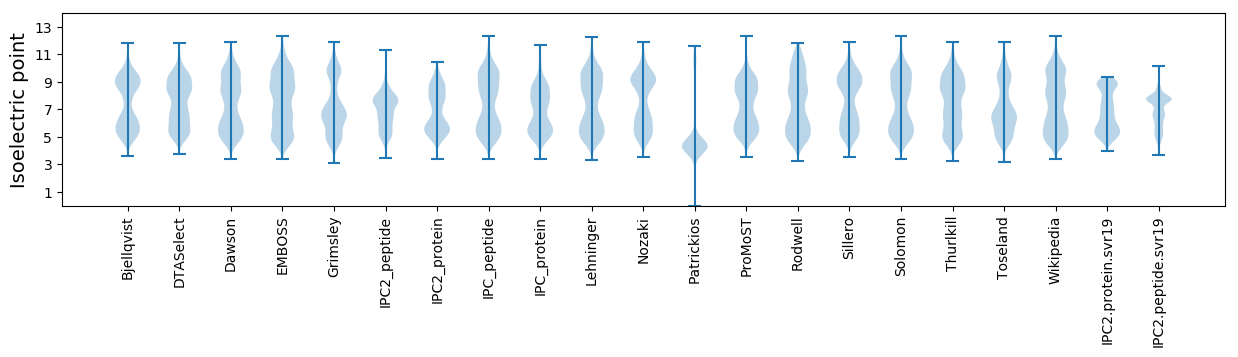
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I7L8P7|I7L8P7_ENCCU Type 1 phosphatases regulator OS=Encephalitozoon cuniculi (strain GB-M1) OX=284813 GN=ECU05_0185 PE=3 SV=1
MM1 pKa = 7.78RR2 pKa = 11.84SINTLIRR9 pKa = 11.84VMLLSEE15 pKa = 4.06ALIYY19 pKa = 10.91ASTEE23 pKa = 3.84GSKK26 pKa = 10.91EE27 pKa = 4.01EE28 pKa = 4.02NSPSITSQQQDD39 pKa = 3.12TSGGEE44 pKa = 3.99EE45 pKa = 3.88EE46 pKa = 4.22NEE48 pKa = 3.91EE49 pKa = 4.07RR50 pKa = 11.84RR51 pKa = 11.84GSGGGEE57 pKa = 3.92EE58 pKa = 4.78APVTAQPQGASGGEE72 pKa = 3.99EE73 pKa = 3.86EE74 pKa = 4.88ADD76 pKa = 3.27GGRR79 pKa = 11.84VVVEE83 pKa = 4.0YY84 pKa = 10.9CSTRR88 pKa = 11.84TQNEE92 pKa = 3.93SGEE95 pKa = 4.53TVEE98 pKa = 4.77TRR100 pKa = 11.84CGCAGDD106 pKa = 5.19CDD108 pKa = 4.07NCCPEE113 pKa = 4.41GMISCLHH120 pKa = 6.0SLGLLL125 pKa = 3.69
MM1 pKa = 7.78RR2 pKa = 11.84SINTLIRR9 pKa = 11.84VMLLSEE15 pKa = 4.06ALIYY19 pKa = 10.91ASTEE23 pKa = 3.84GSKK26 pKa = 10.91EE27 pKa = 4.01EE28 pKa = 4.02NSPSITSQQQDD39 pKa = 3.12TSGGEE44 pKa = 3.99EE45 pKa = 3.88EE46 pKa = 4.22NEE48 pKa = 3.91EE49 pKa = 4.07RR50 pKa = 11.84RR51 pKa = 11.84GSGGGEE57 pKa = 3.92EE58 pKa = 4.78APVTAQPQGASGGEE72 pKa = 3.99EE73 pKa = 3.86EE74 pKa = 4.88ADD76 pKa = 3.27GGRR79 pKa = 11.84VVVEE83 pKa = 4.0YY84 pKa = 10.9CSTRR88 pKa = 11.84TQNEE92 pKa = 3.93SGEE95 pKa = 4.53TVEE98 pKa = 4.77TRR100 pKa = 11.84CGCAGDD106 pKa = 5.19CDD108 pKa = 4.07NCCPEE113 pKa = 4.41GMISCLHH120 pKa = 6.0SLGLLL125 pKa = 3.69
Molecular weight: 13.12 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q8SSA5|Q8SSA5_ENCCU UBIQUITIN CARBOXY-TERMINAL HYDROLASE 14 OS=Encephalitozoon cuniculi (strain GB-M1) OX=284813 GN=ECU03_0660 PE=4 SV=1
MM1 pKa = 7.57GEE3 pKa = 3.81NRR5 pKa = 11.84SRR7 pKa = 11.84RR8 pKa = 11.84TLYY11 pKa = 9.86IRR13 pKa = 11.84NLNRR17 pKa = 11.84RR18 pKa = 11.84LPIRR22 pKa = 11.84EE23 pKa = 3.95VKK25 pKa = 10.1RR26 pKa = 11.84RR27 pKa = 11.84LGLLVSRR34 pKa = 11.84FSGISKK40 pKa = 9.63IKK42 pKa = 9.08MSNRR46 pKa = 11.84PGLLGQAFVHH56 pKa = 6.71LDD58 pKa = 3.53GEE60 pKa = 4.52ITSEE64 pKa = 4.31ILDD67 pKa = 3.71NLEE70 pKa = 3.63GRR72 pKa = 11.84FFLGNVISACPAHH85 pKa = 7.34EE86 pKa = 4.45DD87 pKa = 3.25MCIGRR92 pKa = 11.84RR93 pKa = 11.84RR94 pKa = 11.84STSTRR99 pKa = 11.84TLLVTDD105 pKa = 4.5IPPSVTRR112 pKa = 11.84EE113 pKa = 3.82EE114 pKa = 4.4LEE116 pKa = 3.94GLFRR120 pKa = 11.84GFGGLEE126 pKa = 4.04GIRR129 pKa = 11.84FIRR132 pKa = 11.84VKK134 pKa = 10.81NLAFVDD140 pKa = 4.57FLSSRR145 pKa = 11.84EE146 pKa = 3.71ASAAYY151 pKa = 10.19SHH153 pKa = 7.13FGDD156 pKa = 3.9GLIRR160 pKa = 11.84CGAGEE165 pKa = 4.15MRR167 pKa = 11.84IMPSAQQ173 pKa = 2.84
MM1 pKa = 7.57GEE3 pKa = 3.81NRR5 pKa = 11.84SRR7 pKa = 11.84RR8 pKa = 11.84TLYY11 pKa = 9.86IRR13 pKa = 11.84NLNRR17 pKa = 11.84RR18 pKa = 11.84LPIRR22 pKa = 11.84EE23 pKa = 3.95VKK25 pKa = 10.1RR26 pKa = 11.84RR27 pKa = 11.84LGLLVSRR34 pKa = 11.84FSGISKK40 pKa = 9.63IKK42 pKa = 9.08MSNRR46 pKa = 11.84PGLLGQAFVHH56 pKa = 6.71LDD58 pKa = 3.53GEE60 pKa = 4.52ITSEE64 pKa = 4.31ILDD67 pKa = 3.71NLEE70 pKa = 3.63GRR72 pKa = 11.84FFLGNVISACPAHH85 pKa = 7.34EE86 pKa = 4.45DD87 pKa = 3.25MCIGRR92 pKa = 11.84RR93 pKa = 11.84RR94 pKa = 11.84STSTRR99 pKa = 11.84TLLVTDD105 pKa = 4.5IPPSVTRR112 pKa = 11.84EE113 pKa = 3.82EE114 pKa = 4.4LEE116 pKa = 3.94GLFRR120 pKa = 11.84GFGGLEE126 pKa = 4.04GIRR129 pKa = 11.84FIRR132 pKa = 11.84VKK134 pKa = 10.81NLAFVDD140 pKa = 4.57FLSSRR145 pKa = 11.84EE146 pKa = 3.71ASAAYY151 pKa = 10.19SHH153 pKa = 7.13FGDD156 pKa = 3.9GLIRR160 pKa = 11.84CGAGEE165 pKa = 4.15MRR167 pKa = 11.84IMPSAQQ173 pKa = 2.84
Molecular weight: 19.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
694360 |
25 |
3436 |
340.2 |
38.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.016 ± 0.04 | 2.024 ± 0.025 |
5.55 ± 0.033 | 8.18 ± 0.064 |
4.796 ± 0.044 | 6.491 ± 0.059 |
1.899 ± 0.02 | 6.734 ± 0.045 |
7.157 ± 0.052 | 9.502 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.975 ± 0.024 | 3.942 ± 0.032 |
3.269 ± 0.034 | 2.267 ± 0.03 |
6.674 ± 0.045 | 7.966 ± 0.044 |
4.07 ± 0.033 | 7.073 ± 0.037 |
0.754 ± 0.014 | 3.662 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
