
Lake Sarah-associated circular virus-28
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 6.7
Get precalculated fractions of proteins
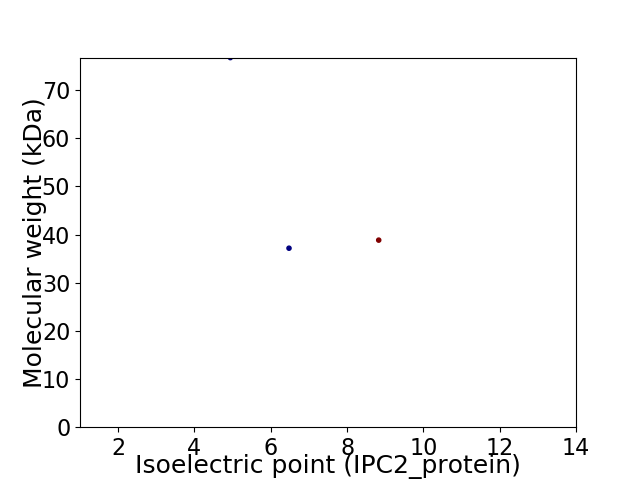
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140AQN0|A0A140AQN0_9VIRU Replication associated protein OS=Lake Sarah-associated circular virus-28 OX=1685755 PE=4 SV=1
MM1 pKa = 7.92ASRR4 pKa = 11.84KK5 pKa = 9.77YY6 pKa = 10.16KK7 pKa = 10.63FCASCFNYY15 pKa = 10.07CDD17 pKa = 3.91SCDD20 pKa = 3.67CLGNLEE26 pKa = 4.17DD27 pKa = 4.84HH28 pKa = 6.03EE29 pKa = 5.29HH30 pKa = 6.57INTFEE35 pKa = 4.25RR36 pKa = 11.84PGKK39 pKa = 10.96GNDD42 pKa = 3.57DD43 pKa = 3.98DD44 pKa = 5.82EE45 pKa = 6.0EE46 pKa = 5.96IDD48 pKa = 5.92DD49 pKa = 5.24EE50 pKa = 5.21EE51 pKa = 5.84EE52 pKa = 4.2DD53 pKa = 4.2DD54 pKa = 4.49NRR56 pKa = 11.84NKK58 pKa = 10.89GGGEE62 pKa = 3.55VWYY65 pKa = 10.0FRR67 pKa = 11.84AQVDD71 pKa = 4.21LTLMAGGNMLLGALMTMEE89 pKa = 4.92EE90 pKa = 4.65MWTDD94 pKa = 3.71IPPDD98 pKa = 3.81NPSLYY103 pKa = 10.75LSIPKK108 pKa = 9.86SRR110 pKa = 11.84FPMDD114 pKa = 3.49PVNIGGSFPNKK125 pKa = 9.53PEE127 pKa = 4.16YY128 pKa = 10.12IKK130 pKa = 10.56PKK132 pKa = 9.64YY133 pKa = 8.52PWEE136 pKa = 3.86IPYY139 pKa = 9.82FQLSYY144 pKa = 10.38PRR146 pKa = 11.84PPEE149 pKa = 4.22KK150 pKa = 10.34SDD152 pKa = 3.53AAIASQAKK160 pKa = 10.2DD161 pKa = 3.52CTKK164 pKa = 10.97DD165 pKa = 3.21CLKK168 pKa = 10.44RR169 pKa = 11.84FSFRR173 pKa = 11.84PVRR176 pKa = 11.84LGICTAKK183 pKa = 10.33CAVIAAGKK191 pKa = 9.87KK192 pKa = 9.83AFNEE196 pKa = 4.31FNVSLEE202 pKa = 4.2DD203 pKa = 3.84IYY205 pKa = 11.54LGVQKK210 pKa = 9.63TINLDD215 pKa = 3.54NLPLYY220 pKa = 10.22IGLIPLIKK228 pKa = 10.23KK229 pKa = 7.08YY230 pKa = 9.44TPPRR234 pKa = 11.84LWNAADD240 pKa = 3.97DD241 pKa = 4.39SVVPGCGFQWVEE253 pKa = 3.46MHH255 pKa = 7.15LIDD258 pKa = 5.74GSDD261 pKa = 3.5PSSDD265 pKa = 3.48FDD267 pKa = 4.65CSGKK271 pKa = 10.51LSSNVDD277 pKa = 3.05GVDD280 pKa = 3.86QFWSDD285 pKa = 3.42NQNGGDD291 pKa = 3.47TSEE294 pKa = 4.28TLKK297 pKa = 10.8DD298 pKa = 3.94YY299 pKa = 11.45YY300 pKa = 10.24PDD302 pKa = 4.23ASDD305 pKa = 3.28PTTDD309 pKa = 2.87VRR311 pKa = 11.84MMMMPHH317 pKa = 6.34VPPPVPPVGPPRR329 pKa = 11.84CTQWALDD336 pKa = 4.06HH337 pKa = 6.49GFCDD341 pKa = 5.39KK342 pKa = 11.38DD343 pKa = 4.21DD344 pKa = 5.17DD345 pKa = 4.63PPPYY349 pKa = 9.99IPPPPTHH356 pKa = 7.09VDD358 pKa = 3.16PPIIVDD364 pKa = 3.75PPIVLPPTGTPPTVLPTYY382 pKa = 10.47DD383 pKa = 2.8ITNNINFPMAVIQYY397 pKa = 9.23NSKK400 pKa = 8.47TRR402 pKa = 11.84ADD404 pKa = 3.66WYY406 pKa = 10.84NSGSTGYY413 pKa = 11.06ADD415 pKa = 4.46DD416 pKa = 3.83RR417 pKa = 11.84QRR419 pKa = 11.84PHH421 pKa = 6.98LNWGAAAIPLKK432 pKa = 10.12TGDD435 pKa = 3.68VIDD438 pKa = 4.52RR439 pKa = 11.84GVTLEE444 pKa = 3.89IPEE447 pKa = 4.17NNYY450 pKa = 10.75EE451 pKa = 4.06YY452 pKa = 11.12VDD454 pKa = 3.57CFFYY458 pKa = 11.36YY459 pKa = 9.49MAHH462 pKa = 6.5RR463 pKa = 11.84PIAGRR468 pKa = 11.84VSMRR472 pKa = 11.84LTYY475 pKa = 10.77VSGPKK480 pKa = 9.59PSGMGFKK487 pKa = 9.88VHH489 pKa = 6.92WDD491 pKa = 3.45NSVKK495 pKa = 10.74PGDD498 pKa = 3.9TQTLRR503 pKa = 11.84GDD505 pKa = 3.32VTTFQFNKK513 pKa = 10.02NMPAGISIHH522 pKa = 5.78SFRR525 pKa = 11.84LTPLHH530 pKa = 6.31VNPPGLQNKK539 pKa = 8.37QEE541 pKa = 4.13FKK543 pKa = 11.44LEE545 pKa = 4.84FFTAWDD551 pKa = 4.0KK552 pKa = 11.33DD553 pKa = 3.97CKK555 pKa = 10.5PVFTLARR562 pKa = 11.84DD563 pKa = 4.1IPWEE567 pKa = 4.14TPYY570 pKa = 11.01GGPPPVFNGIGVVDD584 pKa = 5.33FIGNNNKK591 pKa = 10.07DD592 pKa = 4.29CIAAHH597 pKa = 7.16PIPLNPMVGGVRR609 pKa = 11.84VKK611 pKa = 10.65PPTFSWGNGEE621 pKa = 6.05CIICHH626 pKa = 5.35NQDD629 pKa = 2.99PKK631 pKa = 11.23LPGVAVHH638 pKa = 6.73KK639 pKa = 10.14MNNTGDD645 pKa = 3.64GGITPINSQSYY656 pKa = 7.51FFTWVNPSANGYY668 pKa = 8.27GQGIARR674 pKa = 11.84IDD676 pKa = 3.66GSLGSYY682 pKa = 10.29YY683 pKa = 10.48GACKK687 pKa = 10.26SWTKK691 pKa = 11.09
MM1 pKa = 7.92ASRR4 pKa = 11.84KK5 pKa = 9.77YY6 pKa = 10.16KK7 pKa = 10.63FCASCFNYY15 pKa = 10.07CDD17 pKa = 3.91SCDD20 pKa = 3.67CLGNLEE26 pKa = 4.17DD27 pKa = 4.84HH28 pKa = 6.03EE29 pKa = 5.29HH30 pKa = 6.57INTFEE35 pKa = 4.25RR36 pKa = 11.84PGKK39 pKa = 10.96GNDD42 pKa = 3.57DD43 pKa = 3.98DD44 pKa = 5.82EE45 pKa = 6.0EE46 pKa = 5.96IDD48 pKa = 5.92DD49 pKa = 5.24EE50 pKa = 5.21EE51 pKa = 5.84EE52 pKa = 4.2DD53 pKa = 4.2DD54 pKa = 4.49NRR56 pKa = 11.84NKK58 pKa = 10.89GGGEE62 pKa = 3.55VWYY65 pKa = 10.0FRR67 pKa = 11.84AQVDD71 pKa = 4.21LTLMAGGNMLLGALMTMEE89 pKa = 4.92EE90 pKa = 4.65MWTDD94 pKa = 3.71IPPDD98 pKa = 3.81NPSLYY103 pKa = 10.75LSIPKK108 pKa = 9.86SRR110 pKa = 11.84FPMDD114 pKa = 3.49PVNIGGSFPNKK125 pKa = 9.53PEE127 pKa = 4.16YY128 pKa = 10.12IKK130 pKa = 10.56PKK132 pKa = 9.64YY133 pKa = 8.52PWEE136 pKa = 3.86IPYY139 pKa = 9.82FQLSYY144 pKa = 10.38PRR146 pKa = 11.84PPEE149 pKa = 4.22KK150 pKa = 10.34SDD152 pKa = 3.53AAIASQAKK160 pKa = 10.2DD161 pKa = 3.52CTKK164 pKa = 10.97DD165 pKa = 3.21CLKK168 pKa = 10.44RR169 pKa = 11.84FSFRR173 pKa = 11.84PVRR176 pKa = 11.84LGICTAKK183 pKa = 10.33CAVIAAGKK191 pKa = 9.87KK192 pKa = 9.83AFNEE196 pKa = 4.31FNVSLEE202 pKa = 4.2DD203 pKa = 3.84IYY205 pKa = 11.54LGVQKK210 pKa = 9.63TINLDD215 pKa = 3.54NLPLYY220 pKa = 10.22IGLIPLIKK228 pKa = 10.23KK229 pKa = 7.08YY230 pKa = 9.44TPPRR234 pKa = 11.84LWNAADD240 pKa = 3.97DD241 pKa = 4.39SVVPGCGFQWVEE253 pKa = 3.46MHH255 pKa = 7.15LIDD258 pKa = 5.74GSDD261 pKa = 3.5PSSDD265 pKa = 3.48FDD267 pKa = 4.65CSGKK271 pKa = 10.51LSSNVDD277 pKa = 3.05GVDD280 pKa = 3.86QFWSDD285 pKa = 3.42NQNGGDD291 pKa = 3.47TSEE294 pKa = 4.28TLKK297 pKa = 10.8DD298 pKa = 3.94YY299 pKa = 11.45YY300 pKa = 10.24PDD302 pKa = 4.23ASDD305 pKa = 3.28PTTDD309 pKa = 2.87VRR311 pKa = 11.84MMMMPHH317 pKa = 6.34VPPPVPPVGPPRR329 pKa = 11.84CTQWALDD336 pKa = 4.06HH337 pKa = 6.49GFCDD341 pKa = 5.39KK342 pKa = 11.38DD343 pKa = 4.21DD344 pKa = 5.17DD345 pKa = 4.63PPPYY349 pKa = 9.99IPPPPTHH356 pKa = 7.09VDD358 pKa = 3.16PPIIVDD364 pKa = 3.75PPIVLPPTGTPPTVLPTYY382 pKa = 10.47DD383 pKa = 2.8ITNNINFPMAVIQYY397 pKa = 9.23NSKK400 pKa = 8.47TRR402 pKa = 11.84ADD404 pKa = 3.66WYY406 pKa = 10.84NSGSTGYY413 pKa = 11.06ADD415 pKa = 4.46DD416 pKa = 3.83RR417 pKa = 11.84QRR419 pKa = 11.84PHH421 pKa = 6.98LNWGAAAIPLKK432 pKa = 10.12TGDD435 pKa = 3.68VIDD438 pKa = 4.52RR439 pKa = 11.84GVTLEE444 pKa = 3.89IPEE447 pKa = 4.17NNYY450 pKa = 10.75EE451 pKa = 4.06YY452 pKa = 11.12VDD454 pKa = 3.57CFFYY458 pKa = 11.36YY459 pKa = 9.49MAHH462 pKa = 6.5RR463 pKa = 11.84PIAGRR468 pKa = 11.84VSMRR472 pKa = 11.84LTYY475 pKa = 10.77VSGPKK480 pKa = 9.59PSGMGFKK487 pKa = 9.88VHH489 pKa = 6.92WDD491 pKa = 3.45NSVKK495 pKa = 10.74PGDD498 pKa = 3.9TQTLRR503 pKa = 11.84GDD505 pKa = 3.32VTTFQFNKK513 pKa = 10.02NMPAGISIHH522 pKa = 5.78SFRR525 pKa = 11.84LTPLHH530 pKa = 6.31VNPPGLQNKK539 pKa = 8.37QEE541 pKa = 4.13FKK543 pKa = 11.44LEE545 pKa = 4.84FFTAWDD551 pKa = 4.0KK552 pKa = 11.33DD553 pKa = 3.97CKK555 pKa = 10.5PVFTLARR562 pKa = 11.84DD563 pKa = 4.1IPWEE567 pKa = 4.14TPYY570 pKa = 11.01GGPPPVFNGIGVVDD584 pKa = 5.33FIGNNNKK591 pKa = 10.07DD592 pKa = 4.29CIAAHH597 pKa = 7.16PIPLNPMVGGVRR609 pKa = 11.84VKK611 pKa = 10.65PPTFSWGNGEE621 pKa = 6.05CIICHH626 pKa = 5.35NQDD629 pKa = 2.99PKK631 pKa = 11.23LPGVAVHH638 pKa = 6.73KK639 pKa = 10.14MNNTGDD645 pKa = 3.64GGITPINSQSYY656 pKa = 7.51FFTWVNPSANGYY668 pKa = 8.27GQGIARR674 pKa = 11.84IDD676 pKa = 3.66GSLGSYY682 pKa = 10.29YY683 pKa = 10.48GACKK687 pKa = 10.26SWTKK691 pKa = 11.09
Molecular weight: 76.77 kDa
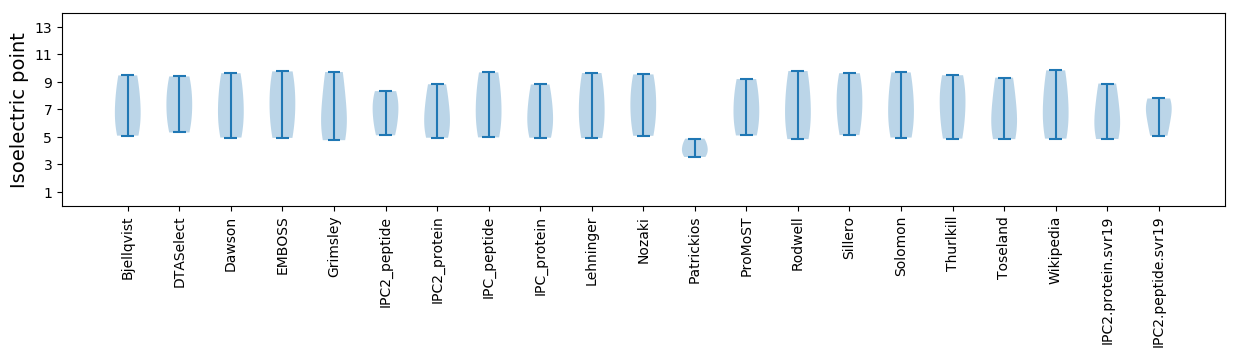
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A126GA16|A0A126GA16_9VIRU Uncharacterized protein OS=Lake Sarah-associated circular virus-28 OX=1685755 PE=4 SV=1
MM1 pKa = 7.27SAVAHH6 pKa = 5.91CAVLRR11 pKa = 11.84NPFSTAVATCKK22 pKa = 10.29IPDD25 pKa = 3.58GSAVLSSSSRR35 pKa = 11.84LSHH38 pKa = 6.26SKK40 pKa = 10.28HH41 pKa = 5.83YY42 pKa = 11.08SCTQNTLCIALYY54 pKa = 10.05PGFQSHH60 pKa = 6.57IFLSGKK66 pKa = 10.02NISQGGGIIGGDD78 pKa = 3.32DD79 pKa = 3.5VANPDD84 pKa = 3.68LQLSSRR90 pKa = 11.84TQKK93 pKa = 10.36EE94 pKa = 4.64GPVGAGGTMTTTQNANAPSKK114 pKa = 10.86YY115 pKa = 10.0RR116 pKa = 11.84IASQGLRR123 pKa = 11.84VSLIEE128 pKa = 4.05NSEE131 pKa = 4.22NNDD134 pKa = 2.55GWFEE138 pKa = 4.72AIRR141 pKa = 11.84IPSAYY146 pKa = 9.73DD147 pKa = 2.9ASDD150 pKa = 3.24FVTLVSTLDD159 pKa = 3.36STSQYY164 pKa = 10.49MGLQKK169 pKa = 10.51DD170 pKa = 3.61KK171 pKa = 11.14KK172 pKa = 10.49IFGGNLGLGTDD183 pKa = 4.03PTSITNWSMNPTYY196 pKa = 9.82ITGKK200 pKa = 9.85LRR202 pKa = 11.84DD203 pKa = 3.85INRR206 pKa = 11.84HH207 pKa = 3.64TFMLHH212 pKa = 6.56RR213 pKa = 11.84EE214 pKa = 3.78NDD216 pKa = 3.15MDD218 pKa = 5.14FVDD221 pKa = 4.15VNNTAEE227 pKa = 4.76FIANGTSVSYY237 pKa = 10.47GKK239 pKa = 10.17SEE241 pKa = 3.87ITRR244 pKa = 11.84AANPSVWWVDD254 pKa = 3.68SNLDD258 pKa = 3.83TILIRR263 pKa = 11.84CFGSIVGSGSGMSVHH278 pKa = 6.08IHH280 pKa = 5.06TVQHH284 pKa = 5.79VEE286 pKa = 4.15EE287 pKa = 4.16QFEE290 pKa = 4.65PNNEE294 pKa = 3.82LARR297 pKa = 11.84YY298 pKa = 7.14MIKK301 pKa = 10.58PPVNTALTTAVMNAIRR317 pKa = 11.84RR318 pKa = 11.84EE319 pKa = 3.99IKK321 pKa = 10.07PSIIRR326 pKa = 11.84APTGGVSVRR335 pKa = 11.84FSRR338 pKa = 11.84TPRR341 pKa = 11.84TYY343 pKa = 10.4RR344 pKa = 11.84RR345 pKa = 11.84RR346 pKa = 11.84KK347 pKa = 7.37SVRR350 pKa = 11.84RR351 pKa = 11.84RR352 pKa = 11.84RR353 pKa = 11.84YY354 pKa = 6.4TT355 pKa = 2.99
MM1 pKa = 7.27SAVAHH6 pKa = 5.91CAVLRR11 pKa = 11.84NPFSTAVATCKK22 pKa = 10.29IPDD25 pKa = 3.58GSAVLSSSSRR35 pKa = 11.84LSHH38 pKa = 6.26SKK40 pKa = 10.28HH41 pKa = 5.83YY42 pKa = 11.08SCTQNTLCIALYY54 pKa = 10.05PGFQSHH60 pKa = 6.57IFLSGKK66 pKa = 10.02NISQGGGIIGGDD78 pKa = 3.32DD79 pKa = 3.5VANPDD84 pKa = 3.68LQLSSRR90 pKa = 11.84TQKK93 pKa = 10.36EE94 pKa = 4.64GPVGAGGTMTTTQNANAPSKK114 pKa = 10.86YY115 pKa = 10.0RR116 pKa = 11.84IASQGLRR123 pKa = 11.84VSLIEE128 pKa = 4.05NSEE131 pKa = 4.22NNDD134 pKa = 2.55GWFEE138 pKa = 4.72AIRR141 pKa = 11.84IPSAYY146 pKa = 9.73DD147 pKa = 2.9ASDD150 pKa = 3.24FVTLVSTLDD159 pKa = 3.36STSQYY164 pKa = 10.49MGLQKK169 pKa = 10.51DD170 pKa = 3.61KK171 pKa = 11.14KK172 pKa = 10.49IFGGNLGLGTDD183 pKa = 4.03PTSITNWSMNPTYY196 pKa = 9.82ITGKK200 pKa = 9.85LRR202 pKa = 11.84DD203 pKa = 3.85INRR206 pKa = 11.84HH207 pKa = 3.64TFMLHH212 pKa = 6.56RR213 pKa = 11.84EE214 pKa = 3.78NDD216 pKa = 3.15MDD218 pKa = 5.14FVDD221 pKa = 4.15VNNTAEE227 pKa = 4.76FIANGTSVSYY237 pKa = 10.47GKK239 pKa = 10.17SEE241 pKa = 3.87ITRR244 pKa = 11.84AANPSVWWVDD254 pKa = 3.68SNLDD258 pKa = 3.83TILIRR263 pKa = 11.84CFGSIVGSGSGMSVHH278 pKa = 6.08IHH280 pKa = 5.06TVQHH284 pKa = 5.79VEE286 pKa = 4.15EE287 pKa = 4.16QFEE290 pKa = 4.65PNNEE294 pKa = 3.82LARR297 pKa = 11.84YY298 pKa = 7.14MIKK301 pKa = 10.58PPVNTALTTAVMNAIRR317 pKa = 11.84RR318 pKa = 11.84EE319 pKa = 3.99IKK321 pKa = 10.07PSIIRR326 pKa = 11.84APTGGVSVRR335 pKa = 11.84FSRR338 pKa = 11.84TPRR341 pKa = 11.84TYY343 pKa = 10.4RR344 pKa = 11.84RR345 pKa = 11.84RR346 pKa = 11.84KK347 pKa = 7.37SVRR350 pKa = 11.84RR351 pKa = 11.84RR352 pKa = 11.84RR353 pKa = 11.84YY354 pKa = 6.4TT355 pKa = 2.99
Molecular weight: 38.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1362 |
316 |
691 |
454.0 |
50.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.947 ± 0.58 | 2.349 ± 0.36 |
7.122 ± 1.043 | 4.772 ± 1.252 |
4.332 ± 0.353 | 7.269 ± 0.888 |
2.129 ± 0.147 | 5.947 ± 0.41 |
5.653 ± 0.954 | 5.653 ± 0.105 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.643 ± 0.07 | 5.727 ± 0.59 |
7.562 ± 2.166 | 2.863 ± 0.27 |
5.36 ± 1.241 | 6.828 ± 1.519 |
5.8 ± 0.982 | 6.094 ± 0.207 |
1.689 ± 0.322 | 4.258 ± 0.686 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
