
Psychroflexus halocasei
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Psychroflexus
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
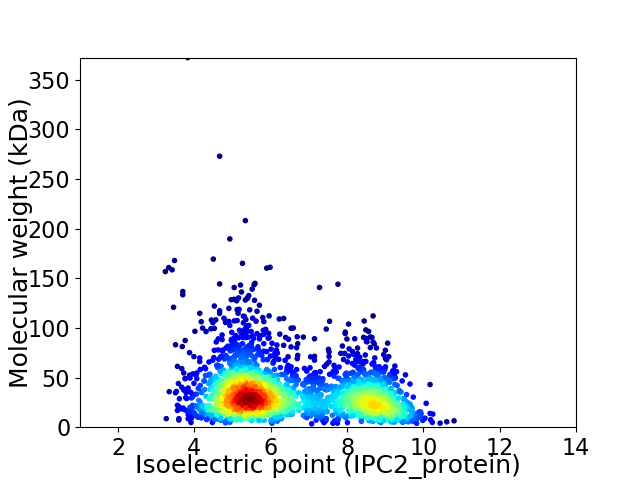
Virtual 2D-PAGE plot for 2462 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H3VQ64|A0A1H3VQ64_9FLAO Pimeloyl-ACP methyl ester carboxylesterase OS=Psychroflexus halocasei OX=908615 GN=SAMN05421540_101203 PE=4 SV=1
MM1 pKa = 7.54KK2 pKa = 10.43NYY4 pKa = 10.2LKK6 pKa = 10.93YY7 pKa = 10.5IGISACAMFLSCEE20 pKa = 4.23PEE22 pKa = 3.63FDD24 pKa = 4.03NQVQEE29 pKa = 3.66NDD31 pKa = 3.34IYY33 pKa = 11.68VSGDD37 pKa = 2.9ADD39 pKa = 3.43FTNFVSVGNSLTAGYY54 pKa = 10.34ADD56 pKa = 3.22NALYY60 pKa = 10.7VQGQEE65 pKa = 3.99SSFPNIMATKK75 pKa = 9.93FEE77 pKa = 4.42LVGGGEE83 pKa = 4.73FKK85 pKa = 10.97QPMMADD91 pKa = 3.21NLGGLLLGGQQISEE105 pKa = 4.07NRR107 pKa = 11.84LVLSVTSGSPAPAILAGTPQTEE129 pKa = 4.26ITNKK133 pKa = 10.39LSGSFNNMGVPGAKK147 pKa = 9.46SFHH150 pKa = 6.36LAAPGYY156 pKa = 9.47GNPAGITSGQANPYY170 pKa = 8.17FARR173 pKa = 11.84FASSEE178 pKa = 4.23SATVIEE184 pKa = 5.37DD185 pKa = 3.55AASQNPTFFSLWIGNNDD202 pKa = 2.98ILSFATSGGIGVDD215 pKa = 3.19QAGNLDD221 pKa = 3.65PSTYY225 pKa = 10.37GSNDD229 pKa = 2.61ITDD232 pKa = 4.16PNVFASVYY240 pKa = 10.66NDD242 pKa = 4.04LLNALNANASGGVVFNIPDD261 pKa = 3.33VTNIPFFTTVPNNALALDD279 pKa = 4.44AAQAQGLTGFFQAFAGIATQTLIQQGVPAAQAQALAAQYY318 pKa = 10.87AITFNEE324 pKa = 4.43GPNRR328 pKa = 11.84FLISVEE334 pKa = 4.14PSQQNPFGFRR344 pKa = 11.84QMTDD348 pKa = 2.95EE349 pKa = 4.54EE350 pKa = 4.96LLLLTIDD357 pKa = 4.86RR358 pKa = 11.84GALQNSGYY366 pKa = 10.71GSVALTPEE374 pKa = 4.14VMQVLGILQQGGTPTPQQAMLVLDD398 pKa = 4.21AVNPIEE404 pKa = 6.27DD405 pKa = 4.34EE406 pKa = 4.37DD407 pKa = 5.13ALDD410 pKa = 4.41DD411 pKa = 4.41DD412 pKa = 5.67EE413 pKa = 6.9IMMVQTAKK421 pKa = 10.72EE422 pKa = 4.27SFNATIEE429 pKa = 4.12GLAQANGLAFVDD441 pKa = 3.54VDD443 pKa = 3.75EE444 pKa = 4.89LMKK447 pKa = 10.66EE448 pKa = 4.22VSTTGIVYY456 pKa = 10.06EE457 pKa = 4.31SGVMTSAFVTGGAFSLDD474 pKa = 3.55GVHH477 pKa = 6.15LTPRR481 pKa = 11.84GYY483 pKa = 11.4AFIASEE489 pKa = 5.06AIQSINEE496 pKa = 4.5TYY498 pKa = 10.71GSTVPTVNIGNFGTVTFSDD517 pKa = 4.3DD518 pKa = 3.46VQQ520 pKa = 4.37
MM1 pKa = 7.54KK2 pKa = 10.43NYY4 pKa = 10.2LKK6 pKa = 10.93YY7 pKa = 10.5IGISACAMFLSCEE20 pKa = 4.23PEE22 pKa = 3.63FDD24 pKa = 4.03NQVQEE29 pKa = 3.66NDD31 pKa = 3.34IYY33 pKa = 11.68VSGDD37 pKa = 2.9ADD39 pKa = 3.43FTNFVSVGNSLTAGYY54 pKa = 10.34ADD56 pKa = 3.22NALYY60 pKa = 10.7VQGQEE65 pKa = 3.99SSFPNIMATKK75 pKa = 9.93FEE77 pKa = 4.42LVGGGEE83 pKa = 4.73FKK85 pKa = 10.97QPMMADD91 pKa = 3.21NLGGLLLGGQQISEE105 pKa = 4.07NRR107 pKa = 11.84LVLSVTSGSPAPAILAGTPQTEE129 pKa = 4.26ITNKK133 pKa = 10.39LSGSFNNMGVPGAKK147 pKa = 9.46SFHH150 pKa = 6.36LAAPGYY156 pKa = 9.47GNPAGITSGQANPYY170 pKa = 8.17FARR173 pKa = 11.84FASSEE178 pKa = 4.23SATVIEE184 pKa = 5.37DD185 pKa = 3.55AASQNPTFFSLWIGNNDD202 pKa = 2.98ILSFATSGGIGVDD215 pKa = 3.19QAGNLDD221 pKa = 3.65PSTYY225 pKa = 10.37GSNDD229 pKa = 2.61ITDD232 pKa = 4.16PNVFASVYY240 pKa = 10.66NDD242 pKa = 4.04LLNALNANASGGVVFNIPDD261 pKa = 3.33VTNIPFFTTVPNNALALDD279 pKa = 4.44AAQAQGLTGFFQAFAGIATQTLIQQGVPAAQAQALAAQYY318 pKa = 10.87AITFNEE324 pKa = 4.43GPNRR328 pKa = 11.84FLISVEE334 pKa = 4.14PSQQNPFGFRR344 pKa = 11.84QMTDD348 pKa = 2.95EE349 pKa = 4.54EE350 pKa = 4.96LLLLTIDD357 pKa = 4.86RR358 pKa = 11.84GALQNSGYY366 pKa = 10.71GSVALTPEE374 pKa = 4.14VMQVLGILQQGGTPTPQQAMLVLDD398 pKa = 4.21AVNPIEE404 pKa = 6.27DD405 pKa = 4.34EE406 pKa = 4.37DD407 pKa = 5.13ALDD410 pKa = 4.41DD411 pKa = 4.41DD412 pKa = 5.67EE413 pKa = 6.9IMMVQTAKK421 pKa = 10.72EE422 pKa = 4.27SFNATIEE429 pKa = 4.12GLAQANGLAFVDD441 pKa = 3.54VDD443 pKa = 3.75EE444 pKa = 4.89LMKK447 pKa = 10.66EE448 pKa = 4.22VSTTGIVYY456 pKa = 10.06EE457 pKa = 4.31SGVMTSAFVTGGAFSLDD474 pKa = 3.55GVHH477 pKa = 6.15LTPRR481 pKa = 11.84GYY483 pKa = 11.4AFIASEE489 pKa = 5.06AIQSINEE496 pKa = 4.5TYY498 pKa = 10.71GSTVPTVNIGNFGTVTFSDD517 pKa = 4.3DD518 pKa = 3.46VQQ520 pKa = 4.37
Molecular weight: 54.64 kDa
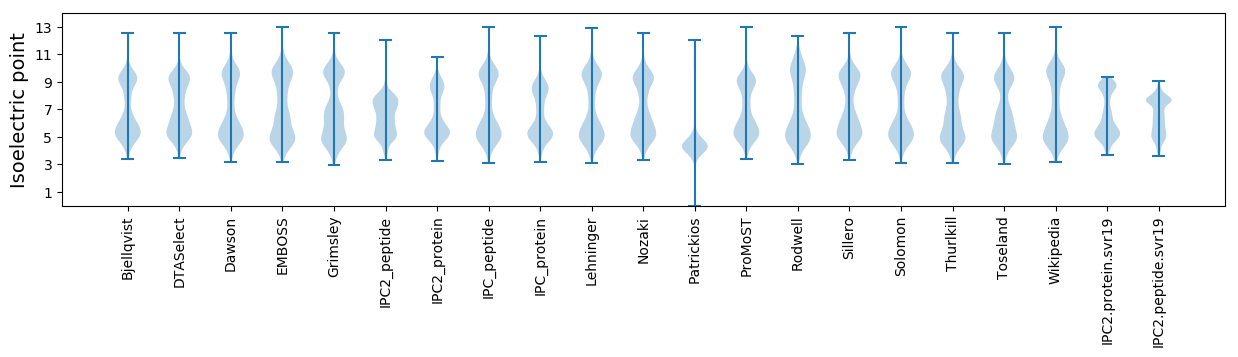
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H3XJQ8|A0A1H3XJQ8_9FLAO Putative fluoride ion transporter CrcB OS=Psychroflexus halocasei OX=908615 GN=crcB PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 10.15RR10 pKa = 11.84KK11 pKa = 9.6RR12 pKa = 11.84KK13 pKa = 8.29NKK15 pKa = 9.34HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.09RR21 pKa = 11.84MASKK25 pKa = 10.5NGRR28 pKa = 11.84KK29 pKa = 9.21VLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.84GRR39 pKa = 11.84KK40 pKa = 7.91RR41 pKa = 11.84LSVSSEE47 pKa = 3.0LSMRR51 pKa = 11.84KK52 pKa = 8.92RR53 pKa = 3.45
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 10.15RR10 pKa = 11.84KK11 pKa = 9.6RR12 pKa = 11.84KK13 pKa = 8.29NKK15 pKa = 9.34HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.09RR21 pKa = 11.84MASKK25 pKa = 10.5NGRR28 pKa = 11.84KK29 pKa = 9.21VLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.84GRR39 pKa = 11.84KK40 pKa = 7.91RR41 pKa = 11.84LSVSSEE47 pKa = 3.0LSMRR51 pKa = 11.84KK52 pKa = 8.92RR53 pKa = 3.45
Molecular weight: 6.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
806779 |
29 |
3454 |
327.7 |
37.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.062 ± 0.038 | 0.668 ± 0.014 |
5.975 ± 0.043 | 7.057 ± 0.047 |
5.339 ± 0.04 | 5.878 ± 0.043 |
1.881 ± 0.021 | 8.084 ± 0.046 |
8.351 ± 0.067 | 9.328 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.218 ± 0.024 | 6.003 ± 0.052 |
3.153 ± 0.024 | 3.829 ± 0.032 |
3.366 ± 0.032 | 6.695 ± 0.039 |
5.207 ± 0.044 | 5.863 ± 0.039 |
0.936 ± 0.016 | 4.105 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
