
Acetobacter sp. DsW_54
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Acetobacteraceae; Acetobacter; unclassified Acetobacter
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
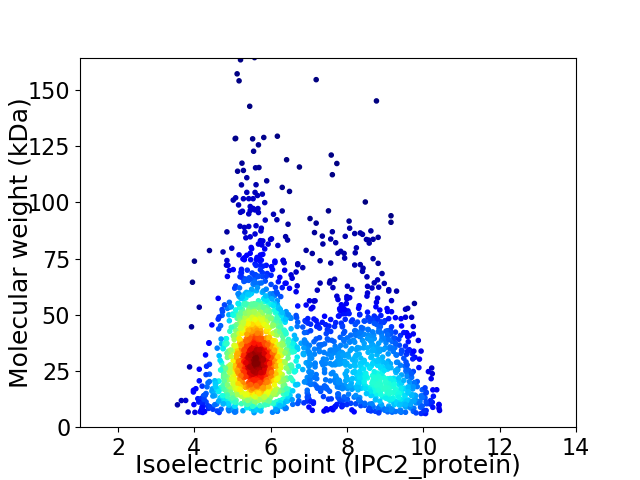
Virtual 2D-PAGE plot for 2200 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A252B1G3|A0A252B1G3_9PROT Glutathione S-transferase OS=Acetobacter sp. DsW_54 OX=1670660 GN=HK20_08005 PE=4 SV=1
MM1 pKa = 7.7SIFNSLTTAVSGINAQSTAFTNLSNNIANSQTVGYY36 pKa = 9.79KK37 pKa = 10.26ADD39 pKa = 3.43TTAFQDD45 pKa = 4.32FVAGAGSTSGSFAQGVANSVAAVTVQHH72 pKa = 6.3VNSQGTASSSTDD84 pKa = 3.09GLAMSISGNGLFNVSKK100 pKa = 9.57KK101 pKa = 9.1TGAVTSAGADD111 pKa = 3.59FASEE115 pKa = 3.86QYY117 pKa = 8.4YY118 pKa = 9.05TRR120 pKa = 11.84NGEE123 pKa = 3.96FYY125 pKa = 10.28EE126 pKa = 5.04DD127 pKa = 3.38KK128 pKa = 10.88DD129 pKa = 4.01GYY131 pKa = 11.26LEE133 pKa = 4.17NTSGYY138 pKa = 9.93YY139 pKa = 10.19LDD141 pKa = 5.32GYY143 pKa = 9.91MVDD146 pKa = 3.67ATTGALDD153 pKa = 3.78TSSVTQINVANVTFRR168 pKa = 11.84PTQTTTITATGTLSSVTGASAVTTNTTVYY197 pKa = 10.83DD198 pKa = 3.58GTYY201 pKa = 10.46NQAKK205 pKa = 9.76PKK207 pKa = 10.82DD208 pKa = 3.68NTGDD212 pKa = 3.47VTLTWTQSSATEE224 pKa = 3.97WTVQASSSTTGMSVTPTTAYY244 pKa = 9.43TVDD247 pKa = 3.98FNTDD251 pKa = 2.86GSLKK255 pKa = 10.66SVTDD259 pKa = 3.45SSGNNVTSTLNGAAANLPISITYY282 pKa = 9.83PDD284 pKa = 4.65GNTQNVNVNIGTIGGTSGMSLSTSSTTPKK313 pKa = 10.38AVSDD317 pKa = 4.03SVTSGTYY324 pKa = 8.99QSASIEE330 pKa = 4.19SDD332 pKa = 3.31GSVMATFDD340 pKa = 3.95NGDD343 pKa = 3.37TQLIGKK349 pKa = 8.62VALSNFANVNGLLAQDD365 pKa = 3.95GQAYY369 pKa = 6.66TATAASGSAKK379 pKa = 9.82TGLVGEE385 pKa = 4.48NATGSLTVGYY395 pKa = 9.77VEE397 pKa = 6.08SSTTDD402 pKa = 3.29LTSDD406 pKa = 3.78LSALIVAQEE415 pKa = 4.3AYY417 pKa = 8.3TANTKK422 pKa = 9.9IVTTADD428 pKa = 3.12QLLQATIAMKK438 pKa = 10.36QQ439 pKa = 3.01
MM1 pKa = 7.7SIFNSLTTAVSGINAQSTAFTNLSNNIANSQTVGYY36 pKa = 9.79KK37 pKa = 10.26ADD39 pKa = 3.43TTAFQDD45 pKa = 4.32FVAGAGSTSGSFAQGVANSVAAVTVQHH72 pKa = 6.3VNSQGTASSSTDD84 pKa = 3.09GLAMSISGNGLFNVSKK100 pKa = 9.57KK101 pKa = 9.1TGAVTSAGADD111 pKa = 3.59FASEE115 pKa = 3.86QYY117 pKa = 8.4YY118 pKa = 9.05TRR120 pKa = 11.84NGEE123 pKa = 3.96FYY125 pKa = 10.28EE126 pKa = 5.04DD127 pKa = 3.38KK128 pKa = 10.88DD129 pKa = 4.01GYY131 pKa = 11.26LEE133 pKa = 4.17NTSGYY138 pKa = 9.93YY139 pKa = 10.19LDD141 pKa = 5.32GYY143 pKa = 9.91MVDD146 pKa = 3.67ATTGALDD153 pKa = 3.78TSSVTQINVANVTFRR168 pKa = 11.84PTQTTTITATGTLSSVTGASAVTTNTTVYY197 pKa = 10.83DD198 pKa = 3.58GTYY201 pKa = 10.46NQAKK205 pKa = 9.76PKK207 pKa = 10.82DD208 pKa = 3.68NTGDD212 pKa = 3.47VTLTWTQSSATEE224 pKa = 3.97WTVQASSSTTGMSVTPTTAYY244 pKa = 9.43TVDD247 pKa = 3.98FNTDD251 pKa = 2.86GSLKK255 pKa = 10.66SVTDD259 pKa = 3.45SSGNNVTSTLNGAAANLPISITYY282 pKa = 9.83PDD284 pKa = 4.65GNTQNVNVNIGTIGGTSGMSLSTSSTTPKK313 pKa = 10.38AVSDD317 pKa = 4.03SVTSGTYY324 pKa = 8.99QSASIEE330 pKa = 4.19SDD332 pKa = 3.31GSVMATFDD340 pKa = 3.95NGDD343 pKa = 3.37TQLIGKK349 pKa = 8.62VALSNFANVNGLLAQDD365 pKa = 3.95GQAYY369 pKa = 6.66TATAASGSAKK379 pKa = 9.82TGLVGEE385 pKa = 4.48NATGSLTVGYY395 pKa = 9.77VEE397 pKa = 6.08SSTTDD402 pKa = 3.29LTSDD406 pKa = 3.78LSALIVAQEE415 pKa = 4.3AYY417 pKa = 8.3TANTKK422 pKa = 9.9IVTTADD428 pKa = 3.12QLLQATIAMKK438 pKa = 10.36QQ439 pKa = 3.01
Molecular weight: 44.64 kDa
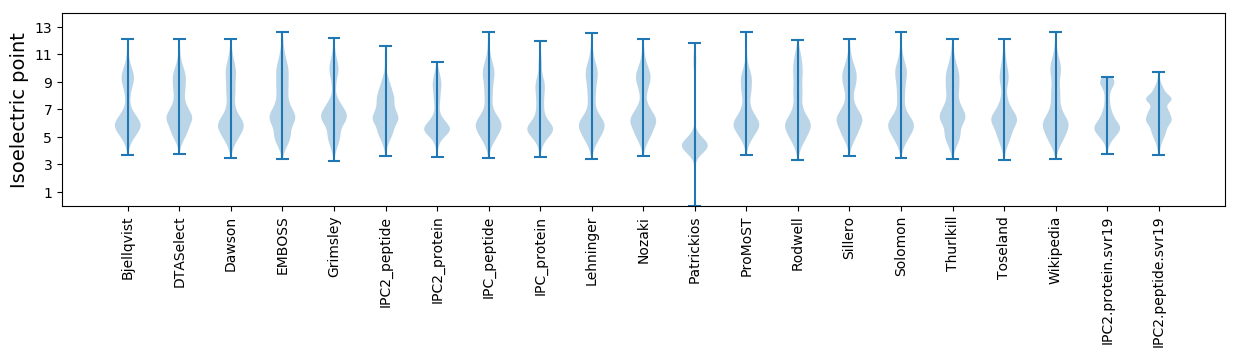
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A252B6A1|A0A252B6A1_9PROT Histidinol phosphate phosphatase OS=Acetobacter sp. DsW_54 OX=1670660 GN=HK20_01425 PE=4 SV=1
MM1 pKa = 7.49KK2 pKa = 10.22RR3 pKa = 11.84RR4 pKa = 11.84LARR7 pKa = 11.84FVEE10 pKa = 4.46LASKK14 pKa = 10.28GRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84HH19 pKa = 4.8VAANDD24 pKa = 3.75TPGVRR29 pKa = 11.84IISWNLLRR37 pKa = 11.84RR38 pKa = 11.84TGATVHH44 pKa = 6.92DD45 pKa = 4.17VAALIEE51 pKa = 4.42AEE53 pKa = 4.18QPDD56 pKa = 4.19ILLMQEE62 pKa = 3.82ATVEE66 pKa = 4.02IDD68 pKa = 3.34VLPDD72 pKa = 3.41VIGGHH77 pKa = 5.71YY78 pKa = 10.16ARR80 pKa = 11.84SPLPGRR86 pKa = 11.84IHH88 pKa = 7.24GVACWSRR95 pKa = 11.84MPFARR100 pKa = 11.84PPRR103 pKa = 11.84ACTIPSGPIVKK114 pKa = 10.03RR115 pKa = 11.84HH116 pKa = 5.16AQIIDD121 pKa = 3.45YY122 pKa = 9.2PQFSLANVHH131 pKa = 6.64LSHH134 pKa = 6.68GQMLNRR140 pKa = 11.84RR141 pKa = 11.84QLRR144 pKa = 11.84RR145 pKa = 11.84IASLLSAPCAILGDD159 pKa = 4.17FNLVGPTLVPGFADD173 pKa = 3.87VGPKK177 pKa = 10.41APTHH181 pKa = 6.29RR182 pKa = 11.84MVDD185 pKa = 3.87LLPIRR190 pKa = 11.84LDD192 pKa = 3.3RR193 pKa = 11.84CLVEE197 pKa = 6.27GMVCSDD203 pKa = 3.5AQVLPVFASDD213 pKa = 3.5HH214 pKa = 6.24RR215 pKa = 11.84PIAVTLQPEE224 pKa = 4.19IGRR227 pKa = 11.84RR228 pKa = 11.84TLASAYY234 pKa = 10.07RR235 pKa = 3.68
MM1 pKa = 7.49KK2 pKa = 10.22RR3 pKa = 11.84RR4 pKa = 11.84LARR7 pKa = 11.84FVEE10 pKa = 4.46LASKK14 pKa = 10.28GRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84HH19 pKa = 4.8VAANDD24 pKa = 3.75TPGVRR29 pKa = 11.84IISWNLLRR37 pKa = 11.84RR38 pKa = 11.84TGATVHH44 pKa = 6.92DD45 pKa = 4.17VAALIEE51 pKa = 4.42AEE53 pKa = 4.18QPDD56 pKa = 4.19ILLMQEE62 pKa = 3.82ATVEE66 pKa = 4.02IDD68 pKa = 3.34VLPDD72 pKa = 3.41VIGGHH77 pKa = 5.71YY78 pKa = 10.16ARR80 pKa = 11.84SPLPGRR86 pKa = 11.84IHH88 pKa = 7.24GVACWSRR95 pKa = 11.84MPFARR100 pKa = 11.84PPRR103 pKa = 11.84ACTIPSGPIVKK114 pKa = 10.03RR115 pKa = 11.84HH116 pKa = 5.16AQIIDD121 pKa = 3.45YY122 pKa = 9.2PQFSLANVHH131 pKa = 6.64LSHH134 pKa = 6.68GQMLNRR140 pKa = 11.84RR141 pKa = 11.84QLRR144 pKa = 11.84RR145 pKa = 11.84IASLLSAPCAILGDD159 pKa = 4.17FNLVGPTLVPGFADD173 pKa = 3.87VGPKK177 pKa = 10.41APTHH181 pKa = 6.29RR182 pKa = 11.84MVDD185 pKa = 3.87LLPIRR190 pKa = 11.84LDD192 pKa = 3.3RR193 pKa = 11.84CLVEE197 pKa = 6.27GMVCSDD203 pKa = 3.5AQVLPVFASDD213 pKa = 3.5HH214 pKa = 6.24RR215 pKa = 11.84PIAVTLQPEE224 pKa = 4.19IGRR227 pKa = 11.84RR228 pKa = 11.84TLASAYY234 pKa = 10.07RR235 pKa = 3.68
Molecular weight: 25.91 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
707855 |
52 |
1555 |
321.8 |
34.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.086 ± 0.067 | 1.087 ± 0.019 |
5.201 ± 0.037 | 5.109 ± 0.062 |
3.497 ± 0.034 | 8.367 ± 0.05 |
2.468 ± 0.023 | 4.706 ± 0.038 |
3.062 ± 0.035 | 10.462 ± 0.063 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.661 ± 0.022 | 2.824 ± 0.034 |
5.546 ± 0.043 | 3.95 ± 0.035 |
6.597 ± 0.05 | 5.512 ± 0.047 |
5.826 ± 0.045 | 7.51 ± 0.047 |
1.325 ± 0.019 | 2.203 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
