
Molossus molossus circovirus 4
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; Circovirus; unclassified Circovirus
Average proteome isoelectric point is 7.98
Get precalculated fractions of proteins
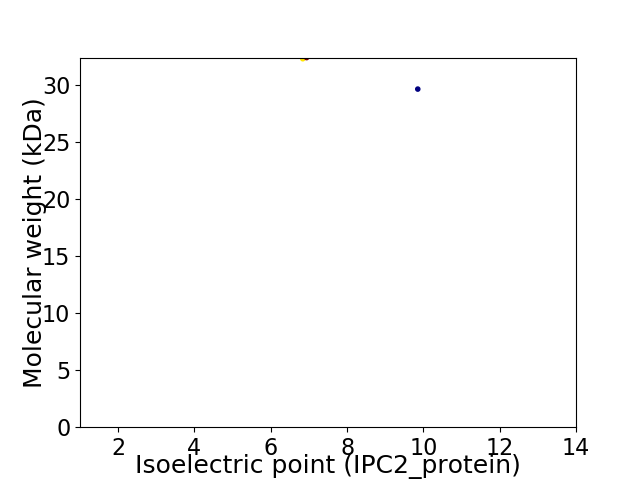
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2I2MP67|A0A2I2MP67_9CIRC Capsid OS=Molossus molossus circovirus 4 OX=1959845 GN=Cap PE=4 SV=1
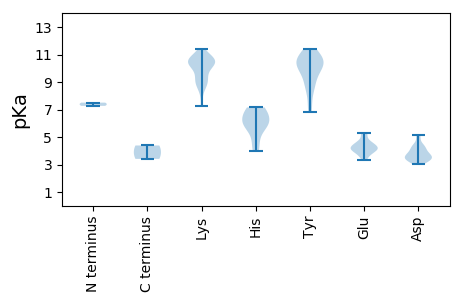
MM1 pKa = 7.27SLSRR5 pKa = 11.84TWCFTSFDD13 pKa = 4.28DD14 pKa = 3.63CWEE17 pKa = 4.1YY18 pKa = 11.39EE19 pKa = 4.54PIQQHH24 pKa = 3.98ITYY27 pKa = 9.37VVWQIEE33 pKa = 4.29KK34 pKa = 10.94CPDD37 pKa = 3.03TGRR40 pKa = 11.84LHH42 pKa = 6.0AQGYY46 pKa = 10.19LEE48 pKa = 4.17TKK50 pKa = 10.56KK51 pKa = 10.44MISHH55 pKa = 6.93KK56 pKa = 8.13EE57 pKa = 3.8AKK59 pKa = 10.32SLIFKK64 pKa = 10.38NNEE67 pKa = 3.49HH68 pKa = 5.92FEE70 pKa = 4.13KK71 pKa = 10.69RR72 pKa = 11.84KK73 pKa = 10.18GSQKK77 pKa = 10.28QAIDD81 pKa = 3.6YY82 pKa = 8.78CKK84 pKa = 10.91KK85 pKa = 10.34NDD87 pKa = 3.48TRR89 pKa = 11.84EE90 pKa = 4.26DD91 pKa = 4.37GPWEE95 pKa = 3.71WGQPTKK101 pKa = 10.49QGQRR105 pKa = 11.84NDD107 pKa = 4.02LIAIKK112 pKa = 10.92NMIDD116 pKa = 3.35EE117 pKa = 4.43GHH119 pKa = 6.77TDD121 pKa = 3.61LEE123 pKa = 4.7VAQEE127 pKa = 4.71HH128 pKa = 5.98FGSWIRR134 pKa = 11.84YY135 pKa = 6.84NKK137 pKa = 10.58SFDD140 pKa = 3.67KK141 pKa = 10.92YY142 pKa = 10.97SKK144 pKa = 9.62LTLRR148 pKa = 11.84CRR150 pKa = 11.84EE151 pKa = 4.03EE152 pKa = 4.29PPTVIILWGPAGCGKK167 pKa = 9.26TSFVYY172 pKa = 10.44KK173 pKa = 9.38KK174 pKa = 9.01TSFVYY179 pKa = 8.56KK180 pKa = 7.29THH182 pKa = 5.78KK183 pKa = 9.13TVYY186 pKa = 10.69SKK188 pKa = 11.4DD189 pKa = 3.32NSKK192 pKa = 8.85WWDD195 pKa = 3.77GYY197 pKa = 10.02HH198 pKa = 4.72QQEE201 pKa = 5.2AILFDD206 pKa = 4.99DD207 pKa = 5.12INWNDD212 pKa = 3.43LDD214 pKa = 3.78RR215 pKa = 11.84RR216 pKa = 11.84EE217 pKa = 4.44ILLLCDD223 pKa = 4.38RR224 pKa = 11.84YY225 pKa = 10.09PLKK228 pKa = 10.99KK229 pKa = 9.59EE230 pKa = 3.93FKK232 pKa = 10.13GGYY235 pKa = 8.59VEE237 pKa = 5.22INSPYY242 pKa = 10.21IYY244 pKa = 9.36FTSNFNPEE252 pKa = 4.07GYY254 pKa = 10.07QIYY257 pKa = 10.7DD258 pKa = 3.44DD259 pKa = 3.82ALKK262 pKa = 10.72RR263 pKa = 11.84RR264 pKa = 11.84INEE267 pKa = 4.43IIPMM271 pKa = 4.39
MM1 pKa = 7.27SLSRR5 pKa = 11.84TWCFTSFDD13 pKa = 4.28DD14 pKa = 3.63CWEE17 pKa = 4.1YY18 pKa = 11.39EE19 pKa = 4.54PIQQHH24 pKa = 3.98ITYY27 pKa = 9.37VVWQIEE33 pKa = 4.29KK34 pKa = 10.94CPDD37 pKa = 3.03TGRR40 pKa = 11.84LHH42 pKa = 6.0AQGYY46 pKa = 10.19LEE48 pKa = 4.17TKK50 pKa = 10.56KK51 pKa = 10.44MISHH55 pKa = 6.93KK56 pKa = 8.13EE57 pKa = 3.8AKK59 pKa = 10.32SLIFKK64 pKa = 10.38NNEE67 pKa = 3.49HH68 pKa = 5.92FEE70 pKa = 4.13KK71 pKa = 10.69RR72 pKa = 11.84KK73 pKa = 10.18GSQKK77 pKa = 10.28QAIDD81 pKa = 3.6YY82 pKa = 8.78CKK84 pKa = 10.91KK85 pKa = 10.34NDD87 pKa = 3.48TRR89 pKa = 11.84EE90 pKa = 4.26DD91 pKa = 4.37GPWEE95 pKa = 3.71WGQPTKK101 pKa = 10.49QGQRR105 pKa = 11.84NDD107 pKa = 4.02LIAIKK112 pKa = 10.92NMIDD116 pKa = 3.35EE117 pKa = 4.43GHH119 pKa = 6.77TDD121 pKa = 3.61LEE123 pKa = 4.7VAQEE127 pKa = 4.71HH128 pKa = 5.98FGSWIRR134 pKa = 11.84YY135 pKa = 6.84NKK137 pKa = 10.58SFDD140 pKa = 3.67KK141 pKa = 10.92YY142 pKa = 10.97SKK144 pKa = 9.62LTLRR148 pKa = 11.84CRR150 pKa = 11.84EE151 pKa = 4.03EE152 pKa = 4.29PPTVIILWGPAGCGKK167 pKa = 9.26TSFVYY172 pKa = 10.44KK173 pKa = 9.38KK174 pKa = 9.01TSFVYY179 pKa = 8.56KK180 pKa = 7.29THH182 pKa = 5.78KK183 pKa = 9.13TVYY186 pKa = 10.69SKK188 pKa = 11.4DD189 pKa = 3.32NSKK192 pKa = 8.85WWDD195 pKa = 3.77GYY197 pKa = 10.02HH198 pKa = 4.72QQEE201 pKa = 5.2AILFDD206 pKa = 4.99DD207 pKa = 5.12INWNDD212 pKa = 3.43LDD214 pKa = 3.78RR215 pKa = 11.84RR216 pKa = 11.84EE217 pKa = 4.44ILLLCDD223 pKa = 4.38RR224 pKa = 11.84YY225 pKa = 10.09PLKK228 pKa = 10.99KK229 pKa = 9.59EE230 pKa = 3.93FKK232 pKa = 10.13GGYY235 pKa = 8.59VEE237 pKa = 5.22INSPYY242 pKa = 10.21IYY244 pKa = 9.36FTSNFNPEE252 pKa = 4.07GYY254 pKa = 10.07QIYY257 pKa = 10.7DD258 pKa = 3.44DD259 pKa = 3.82ALKK262 pKa = 10.72RR263 pKa = 11.84RR264 pKa = 11.84INEE267 pKa = 4.43IIPMM271 pKa = 4.39
Molecular weight: 32.31 kDa
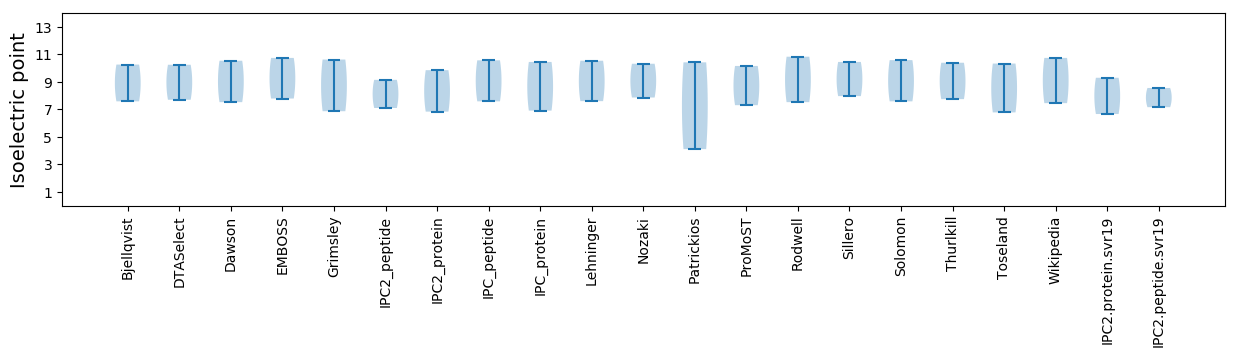
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2I2MP67|A0A2I2MP67_9CIRC Capsid OS=Molossus molossus circovirus 4 OX=1959845 GN=Cap PE=4 SV=1
MM1 pKa = 7.5PFKK4 pKa = 10.49RR5 pKa = 11.84KK6 pKa = 8.71YY7 pKa = 9.75RR8 pKa = 11.84RR9 pKa = 11.84LRR11 pKa = 11.84RR12 pKa = 11.84PAKK15 pKa = 8.8KK16 pKa = 9.47RR17 pKa = 11.84RR18 pKa = 11.84VITRR22 pKa = 11.84PRR24 pKa = 11.84YY25 pKa = 7.16RR26 pKa = 11.84TLARR30 pKa = 11.84GMTANKK36 pKa = 10.27AGIYY40 pKa = 7.97WFKK43 pKa = 10.53RR44 pKa = 11.84TLNASAEE51 pKa = 4.19NGTGISVDD59 pKa = 3.33GTGILQTADD68 pKa = 3.41MFYY71 pKa = 10.23MRR73 pKa = 11.84CNNTAAGSAVFGTMTYY89 pKa = 10.31QFSLQNIPNISEE101 pKa = 4.1FTSLFDD107 pKa = 3.29SYY109 pKa = 10.85QIRR112 pKa = 11.84KK113 pKa = 8.92VVINIIPYY121 pKa = 9.89NNTVQAAQATNVGVAAAYY139 pKa = 9.0WPDD142 pKa = 3.98FSPIMHH148 pKa = 6.12YY149 pKa = 10.84VIDD152 pKa = 4.31HH153 pKa = 7.15DD154 pKa = 4.24GVPPVSANEE163 pKa = 3.72AGINGFQQYY172 pKa = 10.77ASYY175 pKa = 10.52KK176 pKa = 8.65RR177 pKa = 11.84RR178 pKa = 11.84RR179 pKa = 11.84FTGIGPKK186 pKa = 10.2GLSVVIKK193 pKa = 10.23PRR195 pKa = 11.84AKK197 pKa = 10.49LLAGNAALTTVGGTIAGRR215 pKa = 11.84NQWQDD220 pKa = 3.19CAEE223 pKa = 4.06NNLPHH228 pKa = 6.8FGFGMILEE236 pKa = 5.29GISPGTAGAGTIFEE250 pKa = 4.62VPFRR254 pKa = 11.84MEE256 pKa = 3.31TTYY259 pKa = 11.21YY260 pKa = 11.01LKK262 pKa = 10.75FKK264 pKa = 9.87TIRR267 pKa = 3.41
MM1 pKa = 7.5PFKK4 pKa = 10.49RR5 pKa = 11.84KK6 pKa = 8.71YY7 pKa = 9.75RR8 pKa = 11.84RR9 pKa = 11.84LRR11 pKa = 11.84RR12 pKa = 11.84PAKK15 pKa = 8.8KK16 pKa = 9.47RR17 pKa = 11.84RR18 pKa = 11.84VITRR22 pKa = 11.84PRR24 pKa = 11.84YY25 pKa = 7.16RR26 pKa = 11.84TLARR30 pKa = 11.84GMTANKK36 pKa = 10.27AGIYY40 pKa = 7.97WFKK43 pKa = 10.53RR44 pKa = 11.84TLNASAEE51 pKa = 4.19NGTGISVDD59 pKa = 3.33GTGILQTADD68 pKa = 3.41MFYY71 pKa = 10.23MRR73 pKa = 11.84CNNTAAGSAVFGTMTYY89 pKa = 10.31QFSLQNIPNISEE101 pKa = 4.1FTSLFDD107 pKa = 3.29SYY109 pKa = 10.85QIRR112 pKa = 11.84KK113 pKa = 8.92VVINIIPYY121 pKa = 9.89NNTVQAAQATNVGVAAAYY139 pKa = 9.0WPDD142 pKa = 3.98FSPIMHH148 pKa = 6.12YY149 pKa = 10.84VIDD152 pKa = 4.31HH153 pKa = 7.15DD154 pKa = 4.24GVPPVSANEE163 pKa = 3.72AGINGFQQYY172 pKa = 10.77ASYY175 pKa = 10.52KK176 pKa = 8.65RR177 pKa = 11.84RR178 pKa = 11.84RR179 pKa = 11.84FTGIGPKK186 pKa = 10.2GLSVVIKK193 pKa = 10.23PRR195 pKa = 11.84AKK197 pKa = 10.49LLAGNAALTTVGGTIAGRR215 pKa = 11.84NQWQDD220 pKa = 3.19CAEE223 pKa = 4.06NNLPHH228 pKa = 6.8FGFGMILEE236 pKa = 5.29GISPGTAGAGTIFEE250 pKa = 4.62VPFRR254 pKa = 11.84MEE256 pKa = 3.31TTYY259 pKa = 11.21YY260 pKa = 11.01LKK262 pKa = 10.75FKK264 pKa = 9.87TIRR267 pKa = 3.41
Molecular weight: 29.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
538 |
267 |
271 |
269.0 |
30.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.506 ± 2.506 | 1.673 ± 0.642 |
4.833 ± 1.536 | 5.019 ± 1.665 |
5.019 ± 0.416 | 7.435 ± 1.34 |
2.045 ± 0.64 | 7.621 ± 0.09 |
7.435 ± 1.783 | 5.39 ± 0.362 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.23 ± 0.532 | 5.576 ± 0.549 |
4.647 ± 0.415 | 4.275 ± 0.368 |
6.32 ± 1.074 | 5.019 ± 0.364 |
6.877 ± 0.947 | 4.275 ± 0.933 |
2.416 ± 0.898 | 5.39 ± 0.362 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
