
Litoreibacter roseus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Litoreibacter
Average proteome isoelectric point is 5.98
Get precalculated fractions of proteins
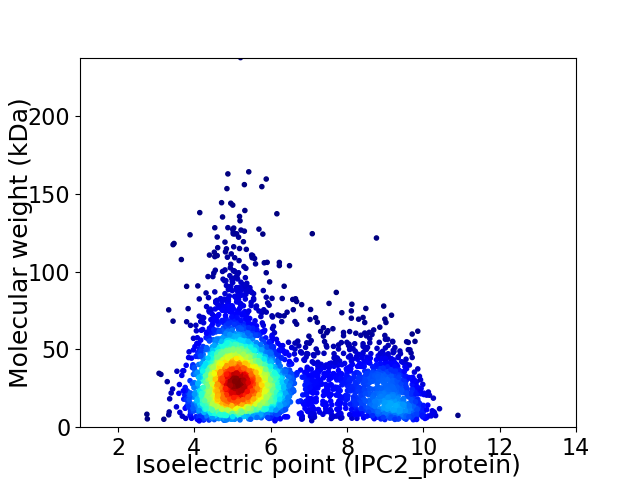
Virtual 2D-PAGE plot for 4462 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6N6JDK3|A0A6N6JDK3_9RHOB Uncharacterized protein OS=Litoreibacter roseus OX=2601869 GN=KIN_12880 PE=4 SV=1
MM1 pKa = 7.23KK2 pKa = 10.13RR3 pKa = 11.84RR4 pKa = 11.84YY5 pKa = 8.69FAGAAALALVAGMAQADD22 pKa = 3.77GHH24 pKa = 5.73QPFAVGEE31 pKa = 4.26GDD33 pKa = 4.79FNWDD37 pKa = 3.11SYY39 pKa = 11.92NAFAEE44 pKa = 4.48KK45 pKa = 10.61YY46 pKa = 8.98DD47 pKa = 4.78LSGQTITITGPWTGLDD63 pKa = 3.6NDD65 pKa = 4.14NVTQVLSYY73 pKa = 10.83FEE75 pKa = 4.58SATGATVNYY84 pKa = 9.53SGSDD88 pKa = 3.17SFEE91 pKa = 3.75QDD93 pKa = 2.48IVISARR99 pKa = 11.84AGSAPNVAVFPQPGLAADD117 pKa = 4.72LASQGLLTPLPEE129 pKa = 4.44GTGDD133 pKa = 3.28WVQSNYY139 pKa = 10.71AAGQSWVDD147 pKa = 3.39LGTYY151 pKa = 9.37PGQDD155 pKa = 3.23GTEE158 pKa = 3.84DD159 pKa = 3.64LYY161 pKa = 11.69GFFYY165 pKa = 10.78KK166 pKa = 10.4VDD168 pKa = 3.56VKK170 pKa = 11.32SLVWYY175 pKa = 10.28SPEE178 pKa = 3.97AFDD181 pKa = 4.71EE182 pKa = 4.48AGYY185 pKa = 10.18DD186 pKa = 3.16IPEE189 pKa = 4.12TMEE192 pKa = 3.89EE193 pKa = 4.3LKK195 pKa = 11.02ALTDD199 pKa = 3.66QIVEE203 pKa = 4.16EE204 pKa = 5.8GGTPWCIGLGSGAATGWPATDD225 pKa = 3.12WVEE228 pKa = 5.29DD229 pKa = 3.51MMLRR233 pKa = 11.84TQSPEE238 pKa = 5.21DD239 pKa = 3.51YY240 pKa = 10.48DD241 pKa = 3.8AWVSNEE247 pKa = 4.14MKK249 pKa = 10.47FDD251 pKa = 4.02DD252 pKa = 4.88PKK254 pKa = 10.76VVAAIEE260 pKa = 4.34EE261 pKa = 4.21YY262 pKa = 10.86GYY264 pKa = 10.28FARR267 pKa = 11.84NNDD270 pKa = 3.71YY271 pKa = 11.43VDD273 pKa = 4.12GGASAVANTDD283 pKa = 3.85FRR285 pKa = 11.84DD286 pKa = 3.78SPTGLFDD293 pKa = 5.06IPPKK297 pKa = 10.57CYY299 pKa = 8.49MHH301 pKa = 6.78RR302 pKa = 11.84QASFIPTFFPEE313 pKa = 4.08DD314 pKa = 3.43AEE316 pKa = 4.3FDD318 pKa = 3.86FFYY321 pKa = 10.97FPAYY325 pKa = 9.54EE326 pKa = 4.4GKK328 pKa = 10.23DD329 pKa = 3.07LGKK332 pKa = 9.53PVLGAGTLWAITNPSDD348 pKa = 3.64GANALMEE355 pKa = 4.1YY356 pKa = 9.93LQTPIAHH363 pKa = 5.68EE364 pKa = 3.88TWMALSGFLTPHH376 pKa = 6.22SGVNTDD382 pKa = 4.32LYY384 pKa = 11.78ANATNKK390 pKa = 10.62ALGDD394 pKa = 3.67VLLNATTFRR403 pKa = 11.84FDD405 pKa = 4.8GSDD408 pKa = 3.54LMPGEE413 pKa = 4.38IGAGAFWTGMVDD425 pKa = 3.36YY426 pKa = 7.62TTGKK430 pKa = 10.29DD431 pKa = 3.13AAEE434 pKa = 4.1VAKK437 pKa = 10.59GIQDD441 pKa = 3.08RR442 pKa = 11.84WDD444 pKa = 3.91TIQQ447 pKa = 4.12
MM1 pKa = 7.23KK2 pKa = 10.13RR3 pKa = 11.84RR4 pKa = 11.84YY5 pKa = 8.69FAGAAALALVAGMAQADD22 pKa = 3.77GHH24 pKa = 5.73QPFAVGEE31 pKa = 4.26GDD33 pKa = 4.79FNWDD37 pKa = 3.11SYY39 pKa = 11.92NAFAEE44 pKa = 4.48KK45 pKa = 10.61YY46 pKa = 8.98DD47 pKa = 4.78LSGQTITITGPWTGLDD63 pKa = 3.6NDD65 pKa = 4.14NVTQVLSYY73 pKa = 10.83FEE75 pKa = 4.58SATGATVNYY84 pKa = 9.53SGSDD88 pKa = 3.17SFEE91 pKa = 3.75QDD93 pKa = 2.48IVISARR99 pKa = 11.84AGSAPNVAVFPQPGLAADD117 pKa = 4.72LASQGLLTPLPEE129 pKa = 4.44GTGDD133 pKa = 3.28WVQSNYY139 pKa = 10.71AAGQSWVDD147 pKa = 3.39LGTYY151 pKa = 9.37PGQDD155 pKa = 3.23GTEE158 pKa = 3.84DD159 pKa = 3.64LYY161 pKa = 11.69GFFYY165 pKa = 10.78KK166 pKa = 10.4VDD168 pKa = 3.56VKK170 pKa = 11.32SLVWYY175 pKa = 10.28SPEE178 pKa = 3.97AFDD181 pKa = 4.71EE182 pKa = 4.48AGYY185 pKa = 10.18DD186 pKa = 3.16IPEE189 pKa = 4.12TMEE192 pKa = 3.89EE193 pKa = 4.3LKK195 pKa = 11.02ALTDD199 pKa = 3.66QIVEE203 pKa = 4.16EE204 pKa = 5.8GGTPWCIGLGSGAATGWPATDD225 pKa = 3.12WVEE228 pKa = 5.29DD229 pKa = 3.51MMLRR233 pKa = 11.84TQSPEE238 pKa = 5.21DD239 pKa = 3.51YY240 pKa = 10.48DD241 pKa = 3.8AWVSNEE247 pKa = 4.14MKK249 pKa = 10.47FDD251 pKa = 4.02DD252 pKa = 4.88PKK254 pKa = 10.76VVAAIEE260 pKa = 4.34EE261 pKa = 4.21YY262 pKa = 10.86GYY264 pKa = 10.28FARR267 pKa = 11.84NNDD270 pKa = 3.71YY271 pKa = 11.43VDD273 pKa = 4.12GGASAVANTDD283 pKa = 3.85FRR285 pKa = 11.84DD286 pKa = 3.78SPTGLFDD293 pKa = 5.06IPPKK297 pKa = 10.57CYY299 pKa = 8.49MHH301 pKa = 6.78RR302 pKa = 11.84QASFIPTFFPEE313 pKa = 4.08DD314 pKa = 3.43AEE316 pKa = 4.3FDD318 pKa = 3.86FFYY321 pKa = 10.97FPAYY325 pKa = 9.54EE326 pKa = 4.4GKK328 pKa = 10.23DD329 pKa = 3.07LGKK332 pKa = 9.53PVLGAGTLWAITNPSDD348 pKa = 3.64GANALMEE355 pKa = 4.1YY356 pKa = 9.93LQTPIAHH363 pKa = 5.68EE364 pKa = 3.88TWMALSGFLTPHH376 pKa = 6.22SGVNTDD382 pKa = 4.32LYY384 pKa = 11.78ANATNKK390 pKa = 10.62ALGDD394 pKa = 3.67VLLNATTFRR403 pKa = 11.84FDD405 pKa = 4.8GSDD408 pKa = 3.54LMPGEE413 pKa = 4.38IGAGAFWTGMVDD425 pKa = 3.36YY426 pKa = 7.62TTGKK430 pKa = 10.29DD431 pKa = 3.13AAEE434 pKa = 4.1VAKK437 pKa = 10.59GIQDD441 pKa = 3.08RR442 pKa = 11.84WDD444 pKa = 3.91TIQQ447 pKa = 4.12
Molecular weight: 48.45 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6N6JEK4|A0A6N6JEK4_9RHOB Uncharacterized protein OS=Litoreibacter roseus OX=2601869 GN=KIN_18550 PE=4 SV=1
MM1 pKa = 7.1NRR3 pKa = 11.84IINMVLRR10 pKa = 11.84QLTRR14 pKa = 11.84QLVRR18 pKa = 11.84RR19 pKa = 11.84GVNSGLNYY27 pKa = 5.4MTKK30 pKa = 10.14RR31 pKa = 11.84GKK33 pKa = 10.39KK34 pKa = 9.07SAAKK38 pKa = 9.84ADD40 pKa = 3.77PSKK43 pKa = 10.52PVPNNAKK50 pKa = 8.93QNRR53 pKa = 11.84RR54 pKa = 11.84AMRR57 pKa = 11.84IMRR60 pKa = 11.84RR61 pKa = 11.84FGRR64 pKa = 11.84FF65 pKa = 2.78
MM1 pKa = 7.1NRR3 pKa = 11.84IINMVLRR10 pKa = 11.84QLTRR14 pKa = 11.84QLVRR18 pKa = 11.84RR19 pKa = 11.84GVNSGLNYY27 pKa = 5.4MTKK30 pKa = 10.14RR31 pKa = 11.84GKK33 pKa = 10.39KK34 pKa = 9.07SAAKK38 pKa = 9.84ADD40 pKa = 3.77PSKK43 pKa = 10.52PVPNNAKK50 pKa = 8.93QNRR53 pKa = 11.84RR54 pKa = 11.84AMRR57 pKa = 11.84IMRR60 pKa = 11.84RR61 pKa = 11.84FGRR64 pKa = 11.84FF65 pKa = 2.78
Molecular weight: 7.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1347393 |
39 |
2131 |
302.0 |
32.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.556 ± 0.043 | 0.9 ± 0.011 |
6.403 ± 0.034 | 5.873 ± 0.036 |
3.928 ± 0.027 | 8.296 ± 0.036 |
2.033 ± 0.021 | 5.504 ± 0.026 |
3.451 ± 0.028 | 9.943 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.719 ± 0.017 | 2.778 ± 0.017 |
4.879 ± 0.025 | 3.291 ± 0.018 |
6.3 ± 0.033 | 5.542 ± 0.025 |
5.763 ± 0.023 | 7.212 ± 0.03 |
1.374 ± 0.016 | 2.253 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
