
Shimia marina
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Shimia
Average proteome isoelectric point is 6.06
Get precalculated fractions of proteins
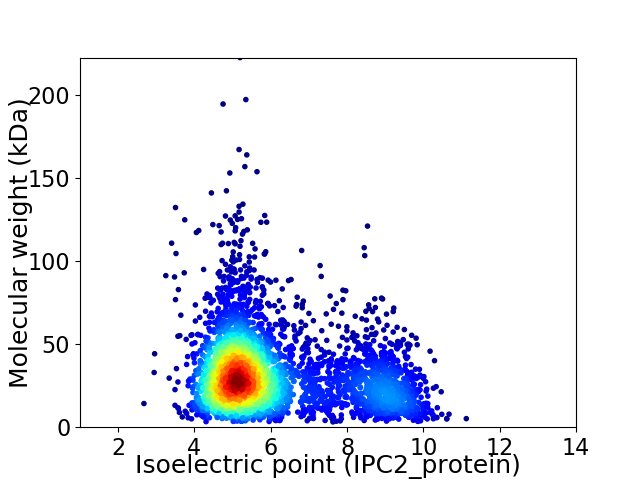
Virtual 2D-PAGE plot for 3874 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
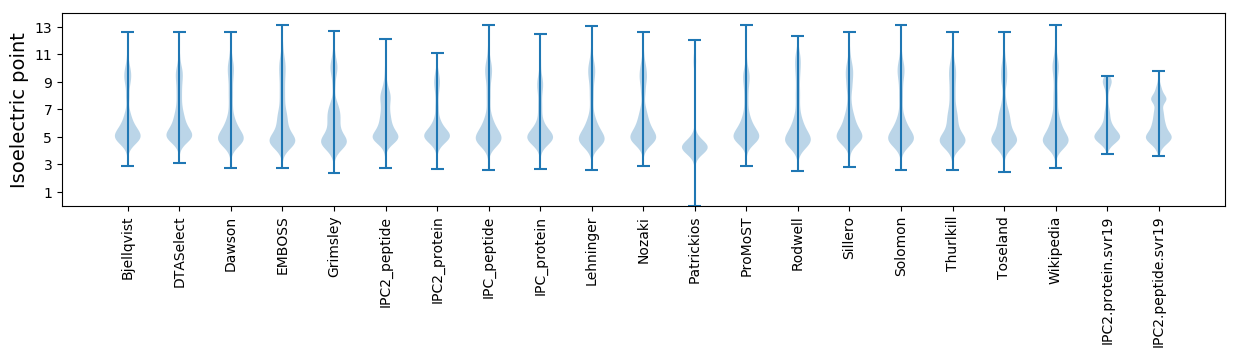
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0N7LRS1|A0A0N7LRS1_9RHOB Ubiquinone/menaquinone biosynthesis C-methyltransferase UbiE OS=Shimia marina OX=321267 GN=ubiE PE=3 SV=1
MM1 pKa = 7.98CIMCAATATFDD12 pKa = 4.15PNRR15 pKa = 11.84HH16 pKa = 5.92PGEE19 pKa = 4.59GPVQALITEE28 pKa = 4.7GNDD31 pKa = 2.79AAAGVGTNYY40 pKa = 9.78TISVGDD46 pKa = 3.89TFNGTLDD53 pKa = 3.49RR54 pKa = 11.84AGDD57 pKa = 3.91RR58 pKa = 11.84DD59 pKa = 3.52WVAIEE64 pKa = 4.02LTADD68 pKa = 2.76TWYY71 pKa = 10.53EE72 pKa = 3.8IGLSGAATSSGTLSDD87 pKa = 4.07PYY89 pKa = 11.1LRR91 pKa = 11.84LYY93 pKa = 10.72DD94 pKa = 3.72SSGNLIGEE102 pKa = 4.38NDD104 pKa = 4.29DD105 pKa = 4.13GGQGYY110 pKa = 10.05EE111 pKa = 4.09SLLGFSASYY120 pKa = 9.78TGTYY124 pKa = 9.74YY125 pKa = 10.15IAAGSYY131 pKa = 10.24RR132 pKa = 11.84DD133 pKa = 4.05GGQGTYY139 pKa = 9.89EE140 pKa = 4.04ISVEE144 pKa = 4.14TAVPGTVDD152 pKa = 3.37EE153 pKa = 4.9LADD156 pKa = 3.88FLTEE160 pKa = 4.23GFWGSEE166 pKa = 4.05RR167 pKa = 11.84HH168 pKa = 5.52WNTSSDD174 pKa = 3.67NIISVNITGLTAAGQQLARR193 pKa = 11.84WAFEE197 pKa = 3.49AWEE200 pKa = 4.22MVANLDD206 pKa = 3.74FQEE209 pKa = 4.37TSGAADD215 pKa = 3.11ITFDD219 pKa = 5.24DD220 pKa = 4.72EE221 pKa = 5.77DD222 pKa = 4.34SGAYY226 pKa = 9.17AQSTVGGGFITSSSVNVGTGWLSSYY251 pKa = 9.12GTTLGSYY258 pKa = 9.87SFQTYY263 pKa = 6.68VHH265 pKa = 7.14EE266 pKa = 4.95IGHH269 pKa = 6.89AIGLGHH275 pKa = 6.24QGGYY279 pKa = 9.86NGSATYY285 pKa = 10.36GVDD288 pKa = 3.12EE289 pKa = 4.63TFSNDD294 pKa = 2.68SWQMSIMSYY303 pKa = 10.51FSQSEE308 pKa = 4.22NTTVDD313 pKa = 2.8ASYY316 pKa = 11.63ARR318 pKa = 11.84LLTAMMADD326 pKa = 3.3IVAAQNLYY334 pKa = 10.38GASTSTDD341 pKa = 2.96GNTVWGSGANLGNYY355 pKa = 9.86LDD357 pKa = 4.8DD358 pKa = 5.35AFAALDD364 pKa = 3.92GSGDD368 pKa = 3.44TSIYY372 pKa = 10.61NGGPVALTIVDD383 pKa = 3.58SGGNDD388 pKa = 3.74LLNLRR393 pKa = 11.84PYY395 pKa = 9.44NTDD398 pKa = 2.44SRR400 pKa = 11.84IDD402 pKa = 3.63LTPGTFSDD410 pKa = 4.18INGLTGNLGIARR422 pKa = 11.84GTVIEE427 pKa = 4.92RR428 pKa = 11.84LTTGNGNDD436 pKa = 3.97TITGNWAGNVVNAGGGNDD454 pKa = 4.21SIDD457 pKa = 4.53GGGGWDD463 pKa = 4.21KK464 pKa = 11.34VWAGDD469 pKa = 4.46GNDD472 pKa = 3.45TVLGGEE478 pKa = 4.54GNDD481 pKa = 3.84TLGGMNGDD489 pKa = 3.57DD490 pKa = 4.47RR491 pKa = 11.84LWGGNGGDD499 pKa = 3.71RR500 pKa = 11.84LYY502 pKa = 11.41GGADD506 pKa = 3.41NDD508 pKa = 4.48TLGGGAGEE516 pKa = 4.5DD517 pKa = 3.35RR518 pKa = 11.84LFGGTGHH525 pKa = 6.97DD526 pKa = 3.56TLRR529 pKa = 11.84GNGDD533 pKa = 3.31SDD535 pKa = 3.81FLRR538 pKa = 11.84GGTGNDD544 pKa = 3.43ALFGDD549 pKa = 5.07DD550 pKa = 5.95GNDD553 pKa = 3.32TLLGDD558 pKa = 4.87DD559 pKa = 4.94GQDD562 pKa = 3.21TLIGGNGADD571 pKa = 3.71SLQAGGWADD580 pKa = 3.64VLEE583 pKa = 4.99GGAGADD589 pKa = 3.89TLLGDD594 pKa = 4.64AGSDD598 pKa = 3.57TLRR601 pKa = 11.84GDD603 pKa = 3.74AGNDD607 pKa = 3.44SLSGGSYY614 pKa = 10.74HH615 pKa = 7.96DD616 pKa = 4.31MLNGGADD623 pKa = 3.49NDD625 pKa = 4.03VLHH628 pKa = 7.29GDD630 pKa = 4.24GGNDD634 pKa = 3.44TLFGGAGNDD643 pKa = 3.49TLYY646 pKa = 11.12GGSGNDD652 pKa = 2.89RR653 pKa = 11.84LFFGAGNDD661 pKa = 3.64VGDD664 pKa = 4.57GGLGADD670 pKa = 3.52TFVFGANVGSDD681 pKa = 3.1NTINNFTVSEE691 pKa = 4.11GDD693 pKa = 3.48RR694 pKa = 11.84LSLDD698 pKa = 3.45DD699 pKa = 5.35ALWLSGHH706 pKa = 5.76GTLSAAEE713 pKa = 4.08VLSTFGSTVGGDD725 pKa = 3.41LVLTFDD731 pKa = 4.67GGQSITLSGLGTTAGLEE748 pKa = 4.16SALDD752 pKa = 3.78LFF754 pKa = 5.27
MM1 pKa = 7.98CIMCAATATFDD12 pKa = 4.15PNRR15 pKa = 11.84HH16 pKa = 5.92PGEE19 pKa = 4.59GPVQALITEE28 pKa = 4.7GNDD31 pKa = 2.79AAAGVGTNYY40 pKa = 9.78TISVGDD46 pKa = 3.89TFNGTLDD53 pKa = 3.49RR54 pKa = 11.84AGDD57 pKa = 3.91RR58 pKa = 11.84DD59 pKa = 3.52WVAIEE64 pKa = 4.02LTADD68 pKa = 2.76TWYY71 pKa = 10.53EE72 pKa = 3.8IGLSGAATSSGTLSDD87 pKa = 4.07PYY89 pKa = 11.1LRR91 pKa = 11.84LYY93 pKa = 10.72DD94 pKa = 3.72SSGNLIGEE102 pKa = 4.38NDD104 pKa = 4.29DD105 pKa = 4.13GGQGYY110 pKa = 10.05EE111 pKa = 4.09SLLGFSASYY120 pKa = 9.78TGTYY124 pKa = 9.74YY125 pKa = 10.15IAAGSYY131 pKa = 10.24RR132 pKa = 11.84DD133 pKa = 4.05GGQGTYY139 pKa = 9.89EE140 pKa = 4.04ISVEE144 pKa = 4.14TAVPGTVDD152 pKa = 3.37EE153 pKa = 4.9LADD156 pKa = 3.88FLTEE160 pKa = 4.23GFWGSEE166 pKa = 4.05RR167 pKa = 11.84HH168 pKa = 5.52WNTSSDD174 pKa = 3.67NIISVNITGLTAAGQQLARR193 pKa = 11.84WAFEE197 pKa = 3.49AWEE200 pKa = 4.22MVANLDD206 pKa = 3.74FQEE209 pKa = 4.37TSGAADD215 pKa = 3.11ITFDD219 pKa = 5.24DD220 pKa = 4.72EE221 pKa = 5.77DD222 pKa = 4.34SGAYY226 pKa = 9.17AQSTVGGGFITSSSVNVGTGWLSSYY251 pKa = 9.12GTTLGSYY258 pKa = 9.87SFQTYY263 pKa = 6.68VHH265 pKa = 7.14EE266 pKa = 4.95IGHH269 pKa = 6.89AIGLGHH275 pKa = 6.24QGGYY279 pKa = 9.86NGSATYY285 pKa = 10.36GVDD288 pKa = 3.12EE289 pKa = 4.63TFSNDD294 pKa = 2.68SWQMSIMSYY303 pKa = 10.51FSQSEE308 pKa = 4.22NTTVDD313 pKa = 2.8ASYY316 pKa = 11.63ARR318 pKa = 11.84LLTAMMADD326 pKa = 3.3IVAAQNLYY334 pKa = 10.38GASTSTDD341 pKa = 2.96GNTVWGSGANLGNYY355 pKa = 9.86LDD357 pKa = 4.8DD358 pKa = 5.35AFAALDD364 pKa = 3.92GSGDD368 pKa = 3.44TSIYY372 pKa = 10.61NGGPVALTIVDD383 pKa = 3.58SGGNDD388 pKa = 3.74LLNLRR393 pKa = 11.84PYY395 pKa = 9.44NTDD398 pKa = 2.44SRR400 pKa = 11.84IDD402 pKa = 3.63LTPGTFSDD410 pKa = 4.18INGLTGNLGIARR422 pKa = 11.84GTVIEE427 pKa = 4.92RR428 pKa = 11.84LTTGNGNDD436 pKa = 3.97TITGNWAGNVVNAGGGNDD454 pKa = 4.21SIDD457 pKa = 4.53GGGGWDD463 pKa = 4.21KK464 pKa = 11.34VWAGDD469 pKa = 4.46GNDD472 pKa = 3.45TVLGGEE478 pKa = 4.54GNDD481 pKa = 3.84TLGGMNGDD489 pKa = 3.57DD490 pKa = 4.47RR491 pKa = 11.84LWGGNGGDD499 pKa = 3.71RR500 pKa = 11.84LYY502 pKa = 11.41GGADD506 pKa = 3.41NDD508 pKa = 4.48TLGGGAGEE516 pKa = 4.5DD517 pKa = 3.35RR518 pKa = 11.84LFGGTGHH525 pKa = 6.97DD526 pKa = 3.56TLRR529 pKa = 11.84GNGDD533 pKa = 3.31SDD535 pKa = 3.81FLRR538 pKa = 11.84GGTGNDD544 pKa = 3.43ALFGDD549 pKa = 5.07DD550 pKa = 5.95GNDD553 pKa = 3.32TLLGDD558 pKa = 4.87DD559 pKa = 4.94GQDD562 pKa = 3.21TLIGGNGADD571 pKa = 3.71SLQAGGWADD580 pKa = 3.64VLEE583 pKa = 4.99GGAGADD589 pKa = 3.89TLLGDD594 pKa = 4.64AGSDD598 pKa = 3.57TLRR601 pKa = 11.84GDD603 pKa = 3.74AGNDD607 pKa = 3.44SLSGGSYY614 pKa = 10.74HH615 pKa = 7.96DD616 pKa = 4.31MLNGGADD623 pKa = 3.49NDD625 pKa = 4.03VLHH628 pKa = 7.29GDD630 pKa = 4.24GGNDD634 pKa = 3.44TLFGGAGNDD643 pKa = 3.49TLYY646 pKa = 11.12GGSGNDD652 pKa = 2.89RR653 pKa = 11.84LFFGAGNDD661 pKa = 3.64VGDD664 pKa = 4.57GGLGADD670 pKa = 3.52TFVFGANVGSDD681 pKa = 3.1NTINNFTVSEE691 pKa = 4.11GDD693 pKa = 3.48RR694 pKa = 11.84LSLDD698 pKa = 3.45DD699 pKa = 5.35ALWLSGHH706 pKa = 5.76GTLSAAEE713 pKa = 4.08VLSTFGSTVGGDD725 pKa = 3.41LVLTFDD731 pKa = 4.67GGQSITLSGLGTTAGLEE748 pKa = 4.16SALDD752 pKa = 3.78LFF754 pKa = 5.27
Molecular weight: 76.83 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0P1EKZ3|A0A0P1EKZ3_9RHOB Putative transposase OrfB OS=Shimia marina OX=321267 GN=SHM7688_00244 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1193212 |
30 |
2003 |
308.0 |
33.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.942 ± 0.053 | 0.907 ± 0.015 |
5.948 ± 0.034 | 6.127 ± 0.038 |
3.86 ± 0.027 | 8.342 ± 0.037 |
2.172 ± 0.021 | 5.139 ± 0.024 |
3.612 ± 0.035 | 9.986 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.886 ± 0.018 | 2.816 ± 0.022 |
4.746 ± 0.029 | 3.557 ± 0.024 |
6.12 ± 0.039 | 5.391 ± 0.029 |
5.54 ± 0.032 | 7.297 ± 0.034 |
1.356 ± 0.016 | 2.257 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
