
Sphingomonas jatrophae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Sphingomonas
Average proteome isoelectric point is 6.72
Get precalculated fractions of proteins
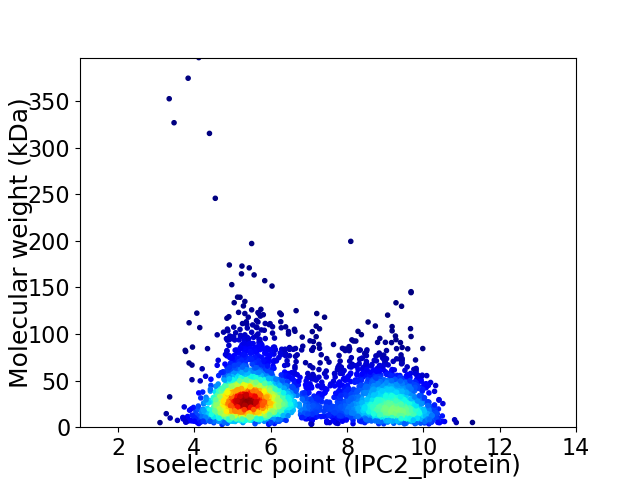
Virtual 2D-PAGE plot for 3810 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I6KZS2|A0A1I6KZS2_9SPHN Uncharacterized protein OS=Sphingomonas jatrophae OX=1166337 GN=SAMN05192580_2064 PE=4 SV=1
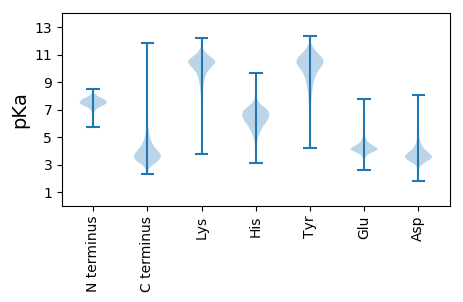
MM1 pKa = 7.85ASPLVRR7 pKa = 11.84FDD9 pKa = 3.94SYY11 pKa = 11.92GSILDD16 pKa = 3.55EE17 pKa = 4.69GAQKK21 pKa = 10.45GRR23 pKa = 11.84PVEE26 pKa = 4.32GFNLTAPPKK35 pKa = 10.05SAAPRR40 pKa = 11.84NVIAADD46 pKa = 3.46ADD48 pKa = 3.85AVVPGSLSGTEE59 pKa = 4.36GYY61 pKa = 10.97DD62 pKa = 3.75LIAVEE67 pKa = 4.94LVAGQTYY74 pKa = 9.98SFAYY78 pKa = 10.1RR79 pKa = 11.84GTPTGGIEE87 pKa = 4.31DD88 pKa = 4.77PYY90 pKa = 11.31LALFDD95 pKa = 4.13TTLSTVLAEE104 pKa = 5.24DD105 pKa = 4.41DD106 pKa = 4.25DD107 pKa = 5.67GGAGRR112 pKa = 11.84SSLITFTAATTGTHH126 pKa = 5.51YY127 pKa = 11.03LYY129 pKa = 10.09ATSFYY134 pKa = 10.56TLDD137 pKa = 3.75TGDD140 pKa = 4.06PSIDD144 pKa = 3.31TGNYY148 pKa = 9.0TIDD151 pKa = 3.08VWSADD156 pKa = 3.78PAKK159 pKa = 10.53DD160 pKa = 3.79APATFAGAEE169 pKa = 4.24LIGAGTTFGFLDD181 pKa = 3.55SAADD185 pKa = 3.37VDD187 pKa = 4.31TYY189 pKa = 11.13RR190 pKa = 11.84IDD192 pKa = 3.69AKK194 pKa = 10.8AGQLYY199 pKa = 8.13TFTYY203 pKa = 10.09AGGIAGSAEE212 pKa = 4.22LADD215 pKa = 5.69PIPGDD220 pKa = 3.85NIGVIDD226 pKa = 3.92IFDD229 pKa = 3.58ARR231 pKa = 11.84GTLVASAVDD240 pKa = 4.1YY241 pKa = 7.46EE242 pKa = 4.51TGASFFAEE250 pKa = 4.11QAGTYY255 pKa = 8.24YY256 pKa = 10.83VQVSPYY262 pKa = 10.53DD263 pKa = 3.55PSLLGGYY270 pKa = 7.37TLDD273 pKa = 3.8VSEE276 pKa = 5.53IDD278 pKa = 3.69PADD281 pKa = 4.14YY282 pKa = 11.12DD283 pKa = 4.15PLDD286 pKa = 4.45SIDD289 pKa = 4.43WRR291 pKa = 11.84DD292 pKa = 3.21ADD294 pKa = 3.73NVPFVDD300 pKa = 3.87VNGVPTAYY308 pKa = 10.54VYY310 pKa = 8.31FAPAGEE316 pKa = 4.44NFGEE320 pKa = 4.35TGDD323 pKa = 4.93DD324 pKa = 3.36GVTPMEE330 pKa = 4.23TFGWQQFQIDD340 pKa = 3.95GVMNALNEE348 pKa = 4.09YY349 pKa = 10.42EE350 pKa = 5.1KK351 pKa = 10.78ILGANYY357 pKa = 9.7EE358 pKa = 4.21ITTDD362 pKa = 3.48PSKK365 pKa = 10.96ATFRR369 pKa = 11.84LLTTEE374 pKa = 4.06SEE376 pKa = 4.27QYY378 pKa = 11.08GAFFYY383 pKa = 10.68PRR385 pKa = 11.84DD386 pKa = 3.69PAYY389 pKa = 9.16GTQQGIGAFNLLSGGFTLPEE409 pKa = 3.97SLQPGGYY416 pKa = 8.52SYY418 pKa = 11.71GVILHH423 pKa = 6.4EE424 pKa = 5.01FGHH427 pKa = 5.85AHH429 pKa = 6.7GLAHH433 pKa = 6.53PHH435 pKa = 6.09DD436 pKa = 4.36TGGGSDD442 pKa = 3.85LMLGVSGPDD451 pKa = 3.12SLGVYY456 pKa = 10.06DD457 pKa = 5.32LNQGVYY463 pKa = 8.91TVMSYY468 pKa = 11.12NDD470 pKa = 3.34GWEE473 pKa = 4.04THH475 pKa = 7.12PDD477 pKa = 3.04GALPFTRR484 pKa = 11.84ATVGYY489 pKa = 8.84GWSGSLSAFDD499 pKa = 3.36IAQLQEE505 pKa = 3.93RR506 pKa = 11.84YY507 pKa = 9.35GVHH510 pKa = 6.19EE511 pKa = 4.59AATGNTTYY519 pKa = 10.89TLDD522 pKa = 4.11DD523 pKa = 3.41VDD525 pKa = 4.11AAGTFYY531 pKa = 10.0QTIWDD536 pKa = 3.82TGGTDD541 pKa = 3.67TIRR544 pKa = 11.84YY545 pKa = 9.38DD546 pKa = 3.43GARR549 pKa = 11.84DD550 pKa = 3.55ARR552 pKa = 11.84IDD554 pKa = 3.82LLAATLDD561 pKa = 3.77YY562 pKa = 10.42TPTGGGVVSYY572 pKa = 11.27VDD574 pKa = 5.44DD575 pKa = 3.28IHH577 pKa = 8.6GGYY580 pKa = 8.83TIANDD585 pKa = 3.65VVIEE589 pKa = 4.03NATGGAGNDD598 pKa = 3.31ALLGNSAKK606 pKa = 10.51NVLTGNAGNDD616 pKa = 3.27ALLGRR621 pKa = 11.84EE622 pKa = 4.38GNDD625 pKa = 2.87RR626 pKa = 11.84LYY628 pKa = 11.33GGAGEE633 pKa = 4.4DD634 pKa = 3.88TLTGGTGLDD643 pKa = 3.98LLFGGAGNDD652 pKa = 3.46VFLMEE657 pKa = 5.47LDD659 pKa = 3.6EE660 pKa = 4.76PANKK664 pKa = 9.92RR665 pKa = 11.84KK666 pKa = 8.44FTLDD670 pKa = 3.42VIGDD674 pKa = 3.93FAKK677 pKa = 10.61GDD679 pKa = 4.07RR680 pKa = 11.84IDD682 pKa = 5.17LSGIDD687 pKa = 3.81ANLNRR692 pKa = 11.84DD693 pKa = 2.99GDD695 pKa = 3.96QAFTMAGKK703 pKa = 9.27FQLLPQIGQIKK714 pKa = 7.66TVSFGSVSAAEE725 pKa = 4.04RR726 pKa = 11.84SLGFDD731 pKa = 4.8LGDD734 pKa = 3.25QKK736 pKa = 11.4AAAGHH741 pKa = 5.78GAVTVLLGEE750 pKa = 4.14VDD752 pKa = 4.63GDD754 pKa = 3.85ILPDD758 pKa = 3.77FAVLMFGSSTVGVSDD773 pKa = 6.13LILL776 pKa = 4.85
MM1 pKa = 7.85ASPLVRR7 pKa = 11.84FDD9 pKa = 3.94SYY11 pKa = 11.92GSILDD16 pKa = 3.55EE17 pKa = 4.69GAQKK21 pKa = 10.45GRR23 pKa = 11.84PVEE26 pKa = 4.32GFNLTAPPKK35 pKa = 10.05SAAPRR40 pKa = 11.84NVIAADD46 pKa = 3.46ADD48 pKa = 3.85AVVPGSLSGTEE59 pKa = 4.36GYY61 pKa = 10.97DD62 pKa = 3.75LIAVEE67 pKa = 4.94LVAGQTYY74 pKa = 9.98SFAYY78 pKa = 10.1RR79 pKa = 11.84GTPTGGIEE87 pKa = 4.31DD88 pKa = 4.77PYY90 pKa = 11.31LALFDD95 pKa = 4.13TTLSTVLAEE104 pKa = 5.24DD105 pKa = 4.41DD106 pKa = 4.25DD107 pKa = 5.67GGAGRR112 pKa = 11.84SSLITFTAATTGTHH126 pKa = 5.51YY127 pKa = 11.03LYY129 pKa = 10.09ATSFYY134 pKa = 10.56TLDD137 pKa = 3.75TGDD140 pKa = 4.06PSIDD144 pKa = 3.31TGNYY148 pKa = 9.0TIDD151 pKa = 3.08VWSADD156 pKa = 3.78PAKK159 pKa = 10.53DD160 pKa = 3.79APATFAGAEE169 pKa = 4.24LIGAGTTFGFLDD181 pKa = 3.55SAADD185 pKa = 3.37VDD187 pKa = 4.31TYY189 pKa = 11.13RR190 pKa = 11.84IDD192 pKa = 3.69AKK194 pKa = 10.8AGQLYY199 pKa = 8.13TFTYY203 pKa = 10.09AGGIAGSAEE212 pKa = 4.22LADD215 pKa = 5.69PIPGDD220 pKa = 3.85NIGVIDD226 pKa = 3.92IFDD229 pKa = 3.58ARR231 pKa = 11.84GTLVASAVDD240 pKa = 4.1YY241 pKa = 7.46EE242 pKa = 4.51TGASFFAEE250 pKa = 4.11QAGTYY255 pKa = 8.24YY256 pKa = 10.83VQVSPYY262 pKa = 10.53DD263 pKa = 3.55PSLLGGYY270 pKa = 7.37TLDD273 pKa = 3.8VSEE276 pKa = 5.53IDD278 pKa = 3.69PADD281 pKa = 4.14YY282 pKa = 11.12DD283 pKa = 4.15PLDD286 pKa = 4.45SIDD289 pKa = 4.43WRR291 pKa = 11.84DD292 pKa = 3.21ADD294 pKa = 3.73NVPFVDD300 pKa = 3.87VNGVPTAYY308 pKa = 10.54VYY310 pKa = 8.31FAPAGEE316 pKa = 4.44NFGEE320 pKa = 4.35TGDD323 pKa = 4.93DD324 pKa = 3.36GVTPMEE330 pKa = 4.23TFGWQQFQIDD340 pKa = 3.95GVMNALNEE348 pKa = 4.09YY349 pKa = 10.42EE350 pKa = 5.1KK351 pKa = 10.78ILGANYY357 pKa = 9.7EE358 pKa = 4.21ITTDD362 pKa = 3.48PSKK365 pKa = 10.96ATFRR369 pKa = 11.84LLTTEE374 pKa = 4.06SEE376 pKa = 4.27QYY378 pKa = 11.08GAFFYY383 pKa = 10.68PRR385 pKa = 11.84DD386 pKa = 3.69PAYY389 pKa = 9.16GTQQGIGAFNLLSGGFTLPEE409 pKa = 3.97SLQPGGYY416 pKa = 8.52SYY418 pKa = 11.71GVILHH423 pKa = 6.4EE424 pKa = 5.01FGHH427 pKa = 5.85AHH429 pKa = 6.7GLAHH433 pKa = 6.53PHH435 pKa = 6.09DD436 pKa = 4.36TGGGSDD442 pKa = 3.85LMLGVSGPDD451 pKa = 3.12SLGVYY456 pKa = 10.06DD457 pKa = 5.32LNQGVYY463 pKa = 8.91TVMSYY468 pKa = 11.12NDD470 pKa = 3.34GWEE473 pKa = 4.04THH475 pKa = 7.12PDD477 pKa = 3.04GALPFTRR484 pKa = 11.84ATVGYY489 pKa = 8.84GWSGSLSAFDD499 pKa = 3.36IAQLQEE505 pKa = 3.93RR506 pKa = 11.84YY507 pKa = 9.35GVHH510 pKa = 6.19EE511 pKa = 4.59AATGNTTYY519 pKa = 10.89TLDD522 pKa = 4.11DD523 pKa = 3.41VDD525 pKa = 4.11AAGTFYY531 pKa = 10.0QTIWDD536 pKa = 3.82TGGTDD541 pKa = 3.67TIRR544 pKa = 11.84YY545 pKa = 9.38DD546 pKa = 3.43GARR549 pKa = 11.84DD550 pKa = 3.55ARR552 pKa = 11.84IDD554 pKa = 3.82LLAATLDD561 pKa = 3.77YY562 pKa = 10.42TPTGGGVVSYY572 pKa = 11.27VDD574 pKa = 5.44DD575 pKa = 3.28IHH577 pKa = 8.6GGYY580 pKa = 8.83TIANDD585 pKa = 3.65VVIEE589 pKa = 4.03NATGGAGNDD598 pKa = 3.31ALLGNSAKK606 pKa = 10.51NVLTGNAGNDD616 pKa = 3.27ALLGRR621 pKa = 11.84EE622 pKa = 4.38GNDD625 pKa = 2.87RR626 pKa = 11.84LYY628 pKa = 11.33GGAGEE633 pKa = 4.4DD634 pKa = 3.88TLTGGTGLDD643 pKa = 3.98LLFGGAGNDD652 pKa = 3.46VFLMEE657 pKa = 5.47LDD659 pKa = 3.6EE660 pKa = 4.76PANKK664 pKa = 9.92RR665 pKa = 11.84KK666 pKa = 8.44FTLDD670 pKa = 3.42VIGDD674 pKa = 3.93FAKK677 pKa = 10.61GDD679 pKa = 4.07RR680 pKa = 11.84IDD682 pKa = 5.17LSGIDD687 pKa = 3.81ANLNRR692 pKa = 11.84DD693 pKa = 2.99GDD695 pKa = 3.96QAFTMAGKK703 pKa = 9.27FQLLPQIGQIKK714 pKa = 7.66TVSFGSVSAAEE725 pKa = 4.04RR726 pKa = 11.84SLGFDD731 pKa = 4.8LGDD734 pKa = 3.25QKK736 pKa = 11.4AAAGHH741 pKa = 5.78GAVTVLLGEE750 pKa = 4.14VDD752 pKa = 4.63GDD754 pKa = 3.85ILPDD758 pKa = 3.77FAVLMFGSSTVGVSDD773 pKa = 6.13LILL776 pKa = 4.85
Molecular weight: 81.29 kDa
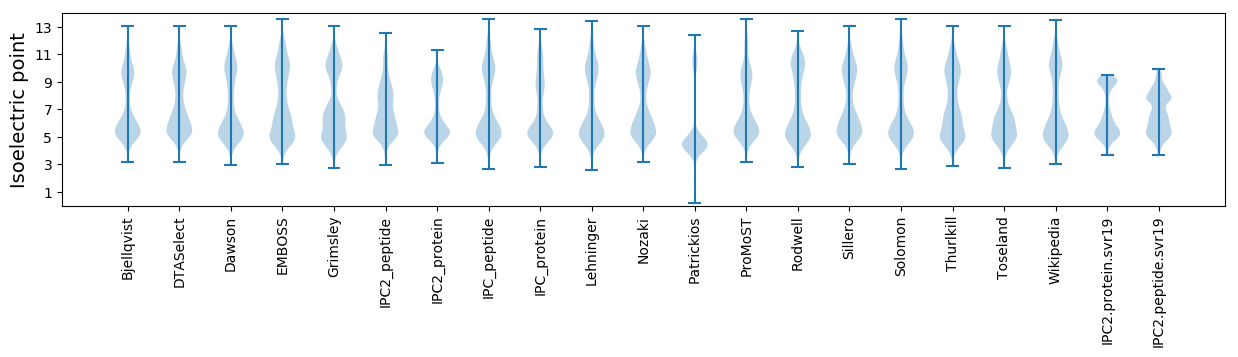
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I6K9R0|A0A1I6K9R0_9SPHN Acetyltransferase (GNAT) domain-containing protein OS=Sphingomonas jatrophae OX=1166337 GN=SAMN05192580_1484 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LVRR12 pKa = 11.84KK13 pKa = 8.99RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.7GFRR19 pKa = 11.84SRR21 pKa = 11.84SATPGGRR28 pKa = 11.84KK29 pKa = 9.04VLAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84NKK41 pKa = 10.45LSAA44 pKa = 3.94
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LVRR12 pKa = 11.84KK13 pKa = 8.99RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.7GFRR19 pKa = 11.84SRR21 pKa = 11.84SATPGGRR28 pKa = 11.84KK29 pKa = 9.04VLAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84NKK41 pKa = 10.45LSAA44 pKa = 3.94
Molecular weight: 5.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1249194 |
29 |
4164 |
327.9 |
35.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.742 ± 0.076 | 0.721 ± 0.013 |
5.915 ± 0.033 | 5.265 ± 0.044 |
3.321 ± 0.029 | 9.265 ± 0.062 |
1.869 ± 0.024 | 4.454 ± 0.025 |
2.44 ± 0.033 | 10.21 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.191 ± 0.023 | 2.199 ± 0.029 |
5.523 ± 0.044 | 2.924 ± 0.023 |
7.984 ± 0.052 | 4.757 ± 0.029 |
5.371 ± 0.053 | 7.358 ± 0.033 |
1.412 ± 0.018 | 2.08 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
