
Burkholderia sp. Leaf177
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Burkholderiaceae; Burkholderia; unclassified Burkholderia
Average proteome isoelectric point is 6.66
Get precalculated fractions of proteins
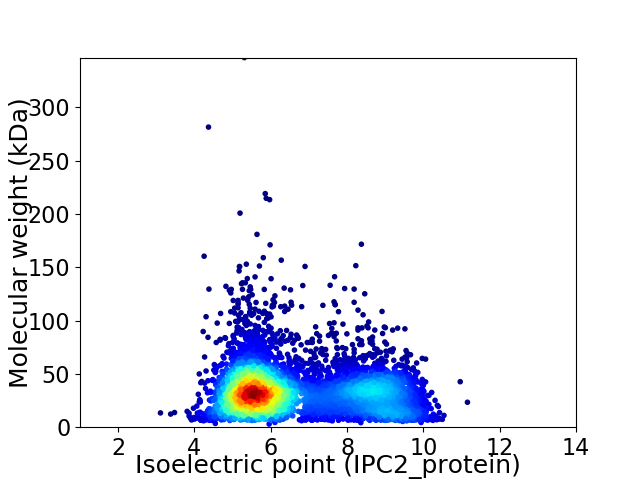
Virtual 2D-PAGE plot for 5955 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
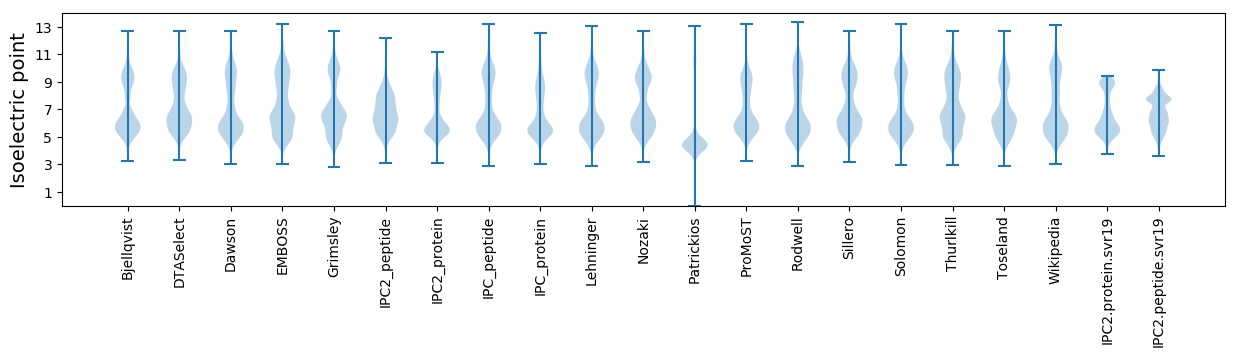
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q5QCS7|A0A0Q5QCS7_9BURK Arsenate reductase OS=Burkholderia sp. Leaf177 OX=1736287 GN=ASG35_03400 PE=3 SV=1
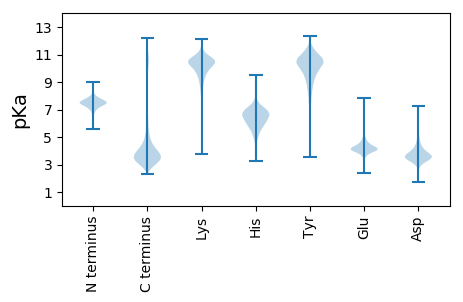
MM1 pKa = 7.9SEE3 pKa = 3.9VLEE6 pKa = 4.24YY7 pKa = 10.77KK8 pKa = 10.32SWVCLICGWIYY19 pKa = 11.0NEE21 pKa = 4.11EE22 pKa = 4.61DD23 pKa = 3.62GLPEE27 pKa = 5.29DD28 pKa = 5.57GIAAGTRR35 pKa = 11.84FADD38 pKa = 4.8IPDD41 pKa = 3.37TWRR44 pKa = 11.84CPLCDD49 pKa = 3.28VGKK52 pKa = 9.89EE53 pKa = 3.91DD54 pKa = 4.28FVAVEE59 pKa = 4.23FF60 pKa = 4.67
MM1 pKa = 7.9SEE3 pKa = 3.9VLEE6 pKa = 4.24YY7 pKa = 10.77KK8 pKa = 10.32SWVCLICGWIYY19 pKa = 11.0NEE21 pKa = 4.11EE22 pKa = 4.61DD23 pKa = 3.62GLPEE27 pKa = 5.29DD28 pKa = 5.57GIAAGTRR35 pKa = 11.84FADD38 pKa = 4.8IPDD41 pKa = 3.37TWRR44 pKa = 11.84CPLCDD49 pKa = 3.28VGKK52 pKa = 9.89EE53 pKa = 3.91DD54 pKa = 4.28FVAVEE59 pKa = 4.23FF60 pKa = 4.67
Molecular weight: 6.8 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q5QAB5|A0A0Q5QAB5_9BURK Ribosome-recycling factor OS=Burkholderia sp. Leaf177 OX=1736287 GN=frr PE=3 SV=1
MM1 pKa = 7.93AIGVHH6 pKa = 5.61HH7 pKa = 6.58VGIVLRR13 pKa = 11.84VLLVLPVLLAVASVVRR29 pKa = 11.84SRR31 pKa = 11.84RR32 pKa = 11.84VTMLQTVIAVRR43 pKa = 11.84GLRR46 pKa = 11.84SAIVAHH52 pKa = 6.52RR53 pKa = 11.84AVIVLRR59 pKa = 11.84DD60 pKa = 3.75PRR62 pKa = 11.84AMASAVRR69 pKa = 11.84SRR71 pKa = 11.84RR72 pKa = 11.84VAMLQEE78 pKa = 4.36VIAVRR83 pKa = 11.84GLRR86 pKa = 11.84LAIVVLRR93 pKa = 11.84AVIVLRR99 pKa = 11.84VLRR102 pKa = 11.84AKK104 pKa = 10.76ASVVRR109 pKa = 11.84SRR111 pKa = 11.84LAVMRR116 pKa = 11.84QGVIGGRR123 pKa = 11.84GLRR126 pKa = 11.84SAIVAHH132 pKa = 6.52RR133 pKa = 11.84AVIVLRR139 pKa = 11.84ILPAMVSVVRR149 pKa = 11.84SRR151 pKa = 11.84RR152 pKa = 11.84VVMRR156 pKa = 11.84QGVNAVHH163 pKa = 7.08GLRR166 pKa = 11.84SAIVAHH172 pKa = 6.52RR173 pKa = 11.84AVIVLRR179 pKa = 11.84VLPAMASVDD188 pKa = 3.17RR189 pKa = 11.84SRR191 pKa = 11.84RR192 pKa = 11.84VVMRR196 pKa = 11.84HH197 pKa = 5.09VMIADD202 pKa = 3.91RR203 pKa = 11.84GLRR206 pKa = 11.84SAIAALHH213 pKa = 4.77VVIVLRR219 pKa = 11.84DD220 pKa = 3.43LQAIANVVHH229 pKa = 6.43SALATTRR236 pKa = 11.84RR237 pKa = 11.84VAQAAIARR245 pKa = 11.84RR246 pKa = 11.84ALLSTTAVVRR256 pKa = 11.84AVRR259 pKa = 11.84APTAAHH265 pKa = 5.9VLLSATAAHH274 pKa = 6.34LVVIAQAVTANAARR288 pKa = 11.84SNRR291 pKa = 11.84GKK293 pKa = 10.38IPVAQEE299 pKa = 3.81AIVRR303 pKa = 11.84RR304 pKa = 11.84VRR306 pKa = 11.84RR307 pKa = 11.84STTTAHH313 pKa = 7.13LAASATTHH321 pKa = 4.74TMAARR326 pKa = 11.84VRR328 pKa = 11.84PLSTANVRR336 pKa = 11.84QATVLTRR343 pKa = 11.84NARR346 pKa = 11.84HH347 pKa = 5.94RR348 pKa = 11.84RR349 pKa = 11.84NASAMSANPHH359 pKa = 6.49ANRR362 pKa = 11.84LQNWQNPATPTHH374 pKa = 6.43HH375 pKa = 6.93RR376 pKa = 11.84HH377 pKa = 6.06PPASPAKK384 pKa = 10.66VKK386 pKa = 10.22MEE388 pKa = 3.68WCAYY392 pKa = 9.91RR393 pKa = 11.84KK394 pKa = 9.8
MM1 pKa = 7.93AIGVHH6 pKa = 5.61HH7 pKa = 6.58VGIVLRR13 pKa = 11.84VLLVLPVLLAVASVVRR29 pKa = 11.84SRR31 pKa = 11.84RR32 pKa = 11.84VTMLQTVIAVRR43 pKa = 11.84GLRR46 pKa = 11.84SAIVAHH52 pKa = 6.52RR53 pKa = 11.84AVIVLRR59 pKa = 11.84DD60 pKa = 3.75PRR62 pKa = 11.84AMASAVRR69 pKa = 11.84SRR71 pKa = 11.84RR72 pKa = 11.84VAMLQEE78 pKa = 4.36VIAVRR83 pKa = 11.84GLRR86 pKa = 11.84LAIVVLRR93 pKa = 11.84AVIVLRR99 pKa = 11.84VLRR102 pKa = 11.84AKK104 pKa = 10.76ASVVRR109 pKa = 11.84SRR111 pKa = 11.84LAVMRR116 pKa = 11.84QGVIGGRR123 pKa = 11.84GLRR126 pKa = 11.84SAIVAHH132 pKa = 6.52RR133 pKa = 11.84AVIVLRR139 pKa = 11.84ILPAMVSVVRR149 pKa = 11.84SRR151 pKa = 11.84RR152 pKa = 11.84VVMRR156 pKa = 11.84QGVNAVHH163 pKa = 7.08GLRR166 pKa = 11.84SAIVAHH172 pKa = 6.52RR173 pKa = 11.84AVIVLRR179 pKa = 11.84VLPAMASVDD188 pKa = 3.17RR189 pKa = 11.84SRR191 pKa = 11.84RR192 pKa = 11.84VVMRR196 pKa = 11.84HH197 pKa = 5.09VMIADD202 pKa = 3.91RR203 pKa = 11.84GLRR206 pKa = 11.84SAIAALHH213 pKa = 4.77VVIVLRR219 pKa = 11.84DD220 pKa = 3.43LQAIANVVHH229 pKa = 6.43SALATTRR236 pKa = 11.84RR237 pKa = 11.84VAQAAIARR245 pKa = 11.84RR246 pKa = 11.84ALLSTTAVVRR256 pKa = 11.84AVRR259 pKa = 11.84APTAAHH265 pKa = 5.9VLLSATAAHH274 pKa = 6.34LVVIAQAVTANAARR288 pKa = 11.84SNRR291 pKa = 11.84GKK293 pKa = 10.38IPVAQEE299 pKa = 3.81AIVRR303 pKa = 11.84RR304 pKa = 11.84VRR306 pKa = 11.84RR307 pKa = 11.84STTTAHH313 pKa = 7.13LAASATTHH321 pKa = 4.74TMAARR326 pKa = 11.84VRR328 pKa = 11.84PLSTANVRR336 pKa = 11.84QATVLTRR343 pKa = 11.84NARR346 pKa = 11.84HH347 pKa = 5.94RR348 pKa = 11.84RR349 pKa = 11.84NASAMSANPHH359 pKa = 6.49ANRR362 pKa = 11.84LQNWQNPATPTHH374 pKa = 6.43HH375 pKa = 6.93RR376 pKa = 11.84HH377 pKa = 6.06PPASPAKK384 pKa = 10.66VKK386 pKa = 10.22MEE388 pKa = 3.68WCAYY392 pKa = 9.91RR393 pKa = 11.84KK394 pKa = 9.8
Molecular weight: 42.64 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1921382 |
24 |
3195 |
322.7 |
35.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.149 ± 0.037 | 0.869 ± 0.011 |
5.495 ± 0.023 | 5.174 ± 0.028 |
3.836 ± 0.023 | 8.037 ± 0.034 |
2.247 ± 0.016 | 5.108 ± 0.02 |
3.53 ± 0.026 | 10.077 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.46 ± 0.016 | 3.074 ± 0.021 |
4.923 ± 0.02 | 3.484 ± 0.019 |
6.494 ± 0.03 | 6.045 ± 0.025 |
5.594 ± 0.023 | 7.707 ± 0.024 |
1.321 ± 0.014 | 2.377 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
