
Ramlibacter sp. H242
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Comamonadaceae; Ramlibacter; unclassified Ramlibacter
Average proteome isoelectric point is 7.49
Get precalculated fractions of proteins
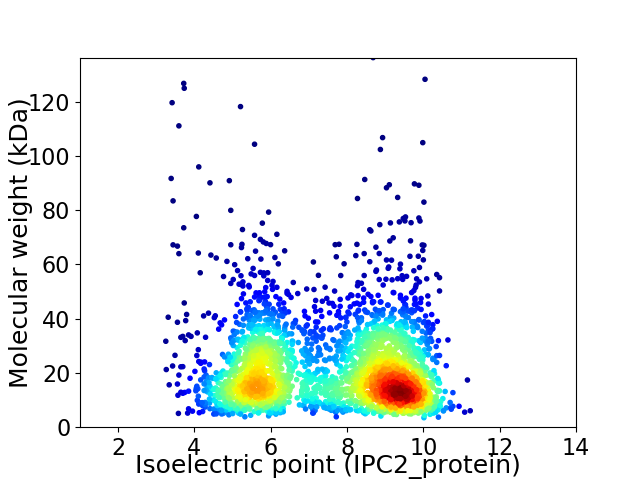
Virtual 2D-PAGE plot for 2805 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6M5XSG0|A0A6M5XSG0_9BURK PhnD/SsuA/transferrin family substrate-binding protein OS=Ramlibacter sp. H242 OX=2732511 GN=HK414_10795 PE=4 SV=1
MM1 pKa = 7.87DD2 pKa = 3.47VHH4 pKa = 7.34AGRR7 pKa = 11.84RR8 pKa = 11.84LPRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84QLHH16 pKa = 4.55GHH18 pKa = 4.57RR19 pKa = 11.84HH20 pKa = 4.34RR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84QRR26 pKa = 11.84RR27 pKa = 11.84HH28 pKa = 5.14PVISLAVTAVADD40 pKa = 4.0IVADD44 pKa = 3.75TASVNEE50 pKa = 4.4DD51 pKa = 2.92ASVTTNLLANDD62 pKa = 3.9SFEE65 pKa = 4.2GAEE68 pKa = 4.29TITAVTQGTHH78 pKa = 4.17GTVTIVDD85 pKa = 3.95PALGTVLYY93 pKa = 10.08TPDD96 pKa = 4.34ANWHH100 pKa = 5.25GTDD103 pKa = 3.06TYY105 pKa = 10.48TYY107 pKa = 8.81TVTSGGVSEE116 pKa = 4.42TATVTVTVAQVNDD129 pKa = 3.67PATFGGATSGSGNEE143 pKa = 4.26DD144 pKa = 3.3DD145 pKa = 5.19AAITGTLTAADD156 pKa = 5.31AIDD159 pKa = 4.62GMTTPAFTVTGAAAHH174 pKa = 5.72GTATINAATGAWSYY188 pKa = 10.91TPAADD193 pKa = 4.23YY194 pKa = 11.36NGTDD198 pKa = 3.15SFTVRR203 pKa = 11.84VTDD206 pKa = 3.32NDD208 pKa = 4.07GNFATQVITLNVASVADD225 pKa = 3.85IAADD229 pKa = 3.54TASVNEE235 pKa = 4.66DD236 pKa = 3.01GSVTSNLLANDD247 pKa = 3.73SFEE250 pKa = 4.2GAEE253 pKa = 4.29TITAVTQGANGTVSIVDD270 pKa = 4.05GAAGTVLYY278 pKa = 9.63TPNANFHH285 pKa = 5.71GTDD288 pKa = 3.06TYY290 pKa = 11.02TYY292 pKa = 8.81TVTSGGVTEE301 pKa = 4.4TATVTVTVAQVDD313 pKa = 4.02DD314 pKa = 3.96PATFGGATSGTGNEE328 pKa = 4.04DD329 pKa = 2.89AAAITGTLTAADD341 pKa = 5.31AIDD344 pKa = 4.62GMTTPAFTVVGAAAHH359 pKa = 6.7GSATINAATGAWSYY373 pKa = 10.91TPAADD378 pKa = 3.86YY379 pKa = 10.75NGPDD383 pKa = 3.31SFTVRR388 pKa = 11.84VTDD391 pKa = 3.81NNGNTATQVISLTVTAVADD410 pKa = 3.94IVADD414 pKa = 3.66TATVNEE420 pKa = 4.57DD421 pKa = 3.58ANVTTNLLANDD432 pKa = 3.9SFEE435 pKa = 4.2GAEE438 pKa = 4.4TITSVTQGTNGTVTILDD455 pKa = 3.81PVLGTVRR462 pKa = 11.84YY463 pKa = 6.97TPNANFHH470 pKa = 5.71GTDD473 pKa = 3.06TYY475 pKa = 11.02TYY477 pKa = 8.81TVTSGGVSEE486 pKa = 4.42TATVTVTVAQVDD498 pKa = 4.02DD499 pKa = 3.96PATFGGATSGSGSEE513 pKa = 4.25DD514 pKa = 2.87AVAITGTLTATDD526 pKa = 5.36AIDD529 pKa = 4.78GMTTPAFTVTGAAAHH544 pKa = 5.72GTATINATTGAWSYY558 pKa = 10.88TPAADD563 pKa = 3.86YY564 pKa = 10.75NGPDD568 pKa = 3.31SFTVRR573 pKa = 11.84VTDD576 pKa = 3.81NNGNTATRR584 pKa = 11.84VISLTVTAVADD595 pKa = 3.92IVADD599 pKa = 3.75TASVNEE605 pKa = 4.41DD606 pKa = 3.17SSVTTNLLTNDD617 pKa = 3.52SFEE620 pKa = 4.21GAEE623 pKa = 4.29TITAVTQGTNGTVTIVNAALGTVLYY648 pKa = 9.59TPNADD653 pKa = 3.33FHH655 pKa = 6.53GTDD658 pKa = 3.06TYY660 pKa = 11.04TYY662 pKa = 8.81TVTSGGVTEE671 pKa = 4.4TATVTVTVAQVDD683 pKa = 4.02DD684 pKa = 3.96PATFGGATTGTGSEE698 pKa = 4.49DD699 pKa = 3.12GAAITGTLTASDD711 pKa = 5.31AIDD714 pKa = 4.12GMTTPAFTVIGNGAHH729 pKa = 7.11GSATINASTGVWSYY743 pKa = 11.23TPVADD748 pKa = 4.1YY749 pKa = 10.96NGPDD753 pKa = 3.33SFTVRR758 pKa = 11.84VTDD761 pKa = 3.81NNGNTATRR769 pKa = 11.84VISLTVTAVADD780 pKa = 3.94IVADD784 pKa = 3.66TATVNEE790 pKa = 4.6DD791 pKa = 3.69SNVTTNLLANDD802 pKa = 4.03TFEE805 pKa = 4.29GAEE808 pKa = 4.25TITAVTQGANGTVTILDD825 pKa = 3.81PVLGTVRR832 pKa = 11.84YY833 pKa = 6.97TPNANFHH840 pKa = 5.71GTDD843 pKa = 3.06TYY845 pKa = 11.02TYY847 pKa = 8.81TVTSGGVSEE856 pKa = 4.42TATVTVTVAQVDD868 pKa = 4.02DD869 pKa = 3.96PATFGGATTGTGNEE883 pKa = 4.18DD884 pKa = 3.26AATITGTLTASDD896 pKa = 5.31AIDD899 pKa = 4.12GMTTPAFTVTGAAAHH914 pKa = 5.72GTATINATTGAWSYY928 pKa = 11.07TPVADD933 pKa = 4.21YY934 pKa = 11.44NGADD938 pKa = 3.39SFTVRR943 pKa = 11.84VTDD946 pKa = 3.81NNGNTATQVITLTVTAAADD965 pKa = 3.56IVADD969 pKa = 3.79TASVNEE975 pKa = 4.41DD976 pKa = 3.17SSVTTNLLTNDD987 pKa = 3.52SFEE990 pKa = 4.21GAEE993 pKa = 4.29TITAVTQGTNGTVTIVNAALGTVLYY1018 pKa = 9.51TPNANFHH1025 pKa = 5.71GTDD1028 pKa = 3.06TYY1030 pKa = 11.02TYY1032 pKa = 8.81TVTSGGVTEE1041 pKa = 4.4TATVTVTVNQVDD1053 pKa = 3.95DD1054 pKa = 3.86PATFGGATTGSGSEE1068 pKa = 4.42DD1069 pKa = 3.11GAAITGTLTASDD1081 pKa = 5.31AIDD1084 pKa = 4.12GMTTPAFTVIGNGAHH1099 pKa = 7.11GSATINASTGVWSYY1113 pKa = 11.23TPVADD1118 pKa = 4.21YY1119 pKa = 11.44NGADD1123 pKa = 3.39SFTVRR1128 pKa = 11.84VTDD1131 pKa = 3.81NNGNTATQVISLSVTAVADD1150 pKa = 3.79IVADD1154 pKa = 3.75TASVNEE1160 pKa = 4.42DD1161 pKa = 3.42ANVTTNLLANDD1172 pKa = 4.03TFEE1175 pKa = 4.29GAEE1178 pKa = 4.25TITAVTQGTNGTVTILDD1195 pKa = 3.81PVLGTVRR1202 pKa = 11.84YY1203 pKa = 6.97TPNANFHH1210 pKa = 5.71GTDD1213 pKa = 3.06TYY1215 pKa = 11.02TYY1217 pKa = 8.81TVTSGGVSEE1226 pKa = 4.42TATVTVTVARR1236 pKa = 11.84STTPPPSAAPRR1247 pKa = 11.84AAAAMKK1253 pKa = 8.34TLPRR1257 pKa = 11.84SRR1259 pKa = 11.84ARR1261 pKa = 3.48
MM1 pKa = 7.87DD2 pKa = 3.47VHH4 pKa = 7.34AGRR7 pKa = 11.84RR8 pKa = 11.84LPRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84QLHH16 pKa = 4.55GHH18 pKa = 4.57RR19 pKa = 11.84HH20 pKa = 4.34RR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84QRR26 pKa = 11.84RR27 pKa = 11.84HH28 pKa = 5.14PVISLAVTAVADD40 pKa = 4.0IVADD44 pKa = 3.75TASVNEE50 pKa = 4.4DD51 pKa = 2.92ASVTTNLLANDD62 pKa = 3.9SFEE65 pKa = 4.2GAEE68 pKa = 4.29TITAVTQGTHH78 pKa = 4.17GTVTIVDD85 pKa = 3.95PALGTVLYY93 pKa = 10.08TPDD96 pKa = 4.34ANWHH100 pKa = 5.25GTDD103 pKa = 3.06TYY105 pKa = 10.48TYY107 pKa = 8.81TVTSGGVSEE116 pKa = 4.42TATVTVTVAQVNDD129 pKa = 3.67PATFGGATSGSGNEE143 pKa = 4.26DD144 pKa = 3.3DD145 pKa = 5.19AAITGTLTAADD156 pKa = 5.31AIDD159 pKa = 4.62GMTTPAFTVTGAAAHH174 pKa = 5.72GTATINAATGAWSYY188 pKa = 10.91TPAADD193 pKa = 4.23YY194 pKa = 11.36NGTDD198 pKa = 3.15SFTVRR203 pKa = 11.84VTDD206 pKa = 3.32NDD208 pKa = 4.07GNFATQVITLNVASVADD225 pKa = 3.85IAADD229 pKa = 3.54TASVNEE235 pKa = 4.66DD236 pKa = 3.01GSVTSNLLANDD247 pKa = 3.73SFEE250 pKa = 4.2GAEE253 pKa = 4.29TITAVTQGANGTVSIVDD270 pKa = 4.05GAAGTVLYY278 pKa = 9.63TPNANFHH285 pKa = 5.71GTDD288 pKa = 3.06TYY290 pKa = 11.02TYY292 pKa = 8.81TVTSGGVTEE301 pKa = 4.4TATVTVTVAQVDD313 pKa = 4.02DD314 pKa = 3.96PATFGGATSGTGNEE328 pKa = 4.04DD329 pKa = 2.89AAAITGTLTAADD341 pKa = 5.31AIDD344 pKa = 4.62GMTTPAFTVVGAAAHH359 pKa = 6.7GSATINAATGAWSYY373 pKa = 10.91TPAADD378 pKa = 3.86YY379 pKa = 10.75NGPDD383 pKa = 3.31SFTVRR388 pKa = 11.84VTDD391 pKa = 3.81NNGNTATQVISLTVTAVADD410 pKa = 3.94IVADD414 pKa = 3.66TATVNEE420 pKa = 4.57DD421 pKa = 3.58ANVTTNLLANDD432 pKa = 3.9SFEE435 pKa = 4.2GAEE438 pKa = 4.4TITSVTQGTNGTVTILDD455 pKa = 3.81PVLGTVRR462 pKa = 11.84YY463 pKa = 6.97TPNANFHH470 pKa = 5.71GTDD473 pKa = 3.06TYY475 pKa = 11.02TYY477 pKa = 8.81TVTSGGVSEE486 pKa = 4.42TATVTVTVAQVDD498 pKa = 4.02DD499 pKa = 3.96PATFGGATSGSGSEE513 pKa = 4.25DD514 pKa = 2.87AVAITGTLTATDD526 pKa = 5.36AIDD529 pKa = 4.78GMTTPAFTVTGAAAHH544 pKa = 5.72GTATINATTGAWSYY558 pKa = 10.88TPAADD563 pKa = 3.86YY564 pKa = 10.75NGPDD568 pKa = 3.31SFTVRR573 pKa = 11.84VTDD576 pKa = 3.81NNGNTATRR584 pKa = 11.84VISLTVTAVADD595 pKa = 3.92IVADD599 pKa = 3.75TASVNEE605 pKa = 4.41DD606 pKa = 3.17SSVTTNLLTNDD617 pKa = 3.52SFEE620 pKa = 4.21GAEE623 pKa = 4.29TITAVTQGTNGTVTIVNAALGTVLYY648 pKa = 9.59TPNADD653 pKa = 3.33FHH655 pKa = 6.53GTDD658 pKa = 3.06TYY660 pKa = 11.04TYY662 pKa = 8.81TVTSGGVTEE671 pKa = 4.4TATVTVTVAQVDD683 pKa = 4.02DD684 pKa = 3.96PATFGGATTGTGSEE698 pKa = 4.49DD699 pKa = 3.12GAAITGTLTASDD711 pKa = 5.31AIDD714 pKa = 4.12GMTTPAFTVIGNGAHH729 pKa = 7.11GSATINASTGVWSYY743 pKa = 11.23TPVADD748 pKa = 4.1YY749 pKa = 10.96NGPDD753 pKa = 3.33SFTVRR758 pKa = 11.84VTDD761 pKa = 3.81NNGNTATRR769 pKa = 11.84VISLTVTAVADD780 pKa = 3.94IVADD784 pKa = 3.66TATVNEE790 pKa = 4.6DD791 pKa = 3.69SNVTTNLLANDD802 pKa = 4.03TFEE805 pKa = 4.29GAEE808 pKa = 4.25TITAVTQGANGTVTILDD825 pKa = 3.81PVLGTVRR832 pKa = 11.84YY833 pKa = 6.97TPNANFHH840 pKa = 5.71GTDD843 pKa = 3.06TYY845 pKa = 11.02TYY847 pKa = 8.81TVTSGGVSEE856 pKa = 4.42TATVTVTVAQVDD868 pKa = 4.02DD869 pKa = 3.96PATFGGATTGTGNEE883 pKa = 4.18DD884 pKa = 3.26AATITGTLTASDD896 pKa = 5.31AIDD899 pKa = 4.12GMTTPAFTVTGAAAHH914 pKa = 5.72GTATINATTGAWSYY928 pKa = 11.07TPVADD933 pKa = 4.21YY934 pKa = 11.44NGADD938 pKa = 3.39SFTVRR943 pKa = 11.84VTDD946 pKa = 3.81NNGNTATQVITLTVTAAADD965 pKa = 3.56IVADD969 pKa = 3.79TASVNEE975 pKa = 4.41DD976 pKa = 3.17SSVTTNLLTNDD987 pKa = 3.52SFEE990 pKa = 4.21GAEE993 pKa = 4.29TITAVTQGTNGTVTIVNAALGTVLYY1018 pKa = 9.51TPNANFHH1025 pKa = 5.71GTDD1028 pKa = 3.06TYY1030 pKa = 11.02TYY1032 pKa = 8.81TVTSGGVTEE1041 pKa = 4.4TATVTVTVNQVDD1053 pKa = 3.95DD1054 pKa = 3.86PATFGGATTGSGSEE1068 pKa = 4.42DD1069 pKa = 3.11GAAITGTLTASDD1081 pKa = 5.31AIDD1084 pKa = 4.12GMTTPAFTVIGNGAHH1099 pKa = 7.11GSATINASTGVWSYY1113 pKa = 11.23TPVADD1118 pKa = 4.21YY1119 pKa = 11.44NGADD1123 pKa = 3.39SFTVRR1128 pKa = 11.84VTDD1131 pKa = 3.81NNGNTATQVISLSVTAVADD1150 pKa = 3.79IVADD1154 pKa = 3.75TASVNEE1160 pKa = 4.42DD1161 pKa = 3.42ANVTTNLLANDD1172 pKa = 4.03TFEE1175 pKa = 4.29GAEE1178 pKa = 4.25TITAVTQGTNGTVTILDD1195 pKa = 3.81PVLGTVRR1202 pKa = 11.84YY1203 pKa = 6.97TPNANFHH1210 pKa = 5.71GTDD1213 pKa = 3.06TYY1215 pKa = 11.02TYY1217 pKa = 8.81TVTSGGVSEE1226 pKa = 4.42TATVTVTVARR1236 pKa = 11.84STTPPPSAAPRR1247 pKa = 11.84AAAAMKK1253 pKa = 8.34TLPRR1257 pKa = 11.84SRR1259 pKa = 11.84ARR1261 pKa = 3.48
Molecular weight: 126.71 kDa
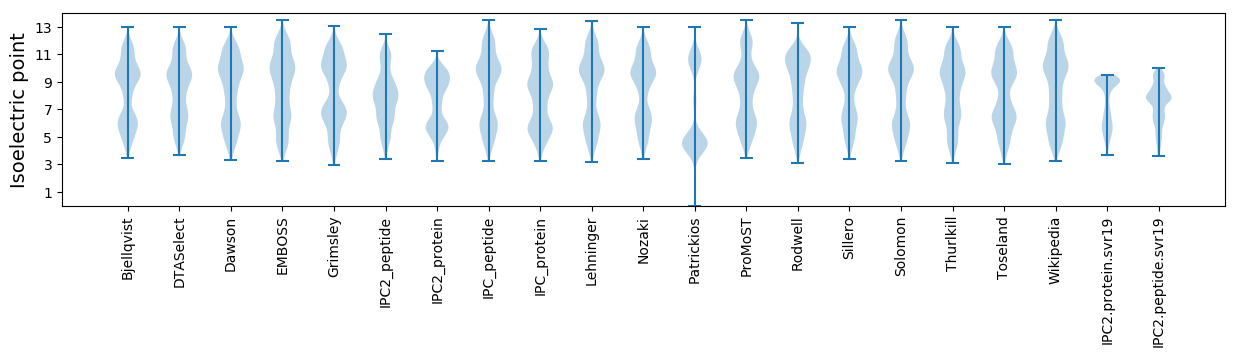
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6M5XVB6|A0A6M5XVB6_9BURK Uncharacterized protein OS=Ramlibacter sp. H242 OX=2732511 GN=HK414_19700 PE=4 SV=1
MM1 pKa = 6.84QRR3 pKa = 11.84KK4 pKa = 6.25TFHH7 pKa = 6.85RR8 pKa = 11.84FLLAAAAAATLGWGSARR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84SPRR30 pKa = 11.84FRR32 pKa = 11.84SSTRR36 pKa = 11.84WPSAARR42 pKa = 11.84SRR44 pKa = 11.84RR45 pKa = 11.84PSTAMPPTSS54 pKa = 3.31
MM1 pKa = 6.84QRR3 pKa = 11.84KK4 pKa = 6.25TFHH7 pKa = 6.85RR8 pKa = 11.84FLLAAAAAATLGWGSARR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84SPRR30 pKa = 11.84FRR32 pKa = 11.84SSTRR36 pKa = 11.84WPSAARR42 pKa = 11.84SRR44 pKa = 11.84RR45 pKa = 11.84PSTAMPPTSS54 pKa = 3.31
Molecular weight: 6.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
608434 |
33 |
1265 |
216.9 |
23.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.802 ± 0.072 | 1.162 ± 0.019 |
5.07 ± 0.044 | 4.964 ± 0.046 |
3.136 ± 0.031 | 8.938 ± 0.065 |
2.243 ± 0.029 | 3.557 ± 0.036 |
2.724 ± 0.044 | 9.537 ± 0.067 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.371 ± 0.027 | 2.338 ± 0.031 |
5.969 ± 0.049 | 3.489 ± 0.037 |
8.571 ± 0.076 | 5.542 ± 0.04 |
5.631 ± 0.092 | 7.641 ± 0.045 |
1.426 ± 0.024 | 1.889 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
