
Pepper cryptic virus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; Deltapartitivirus
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
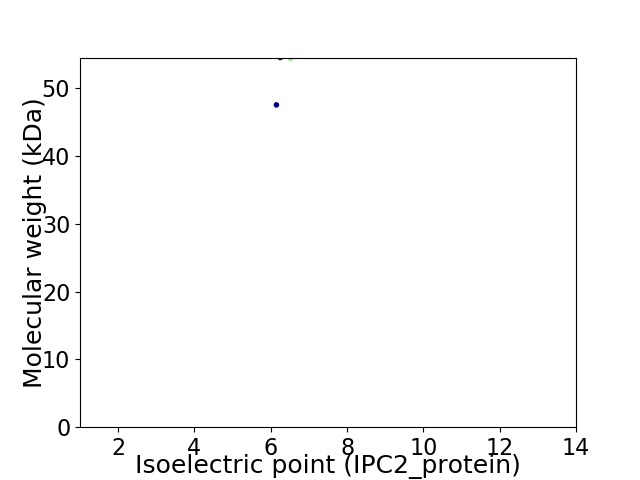
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
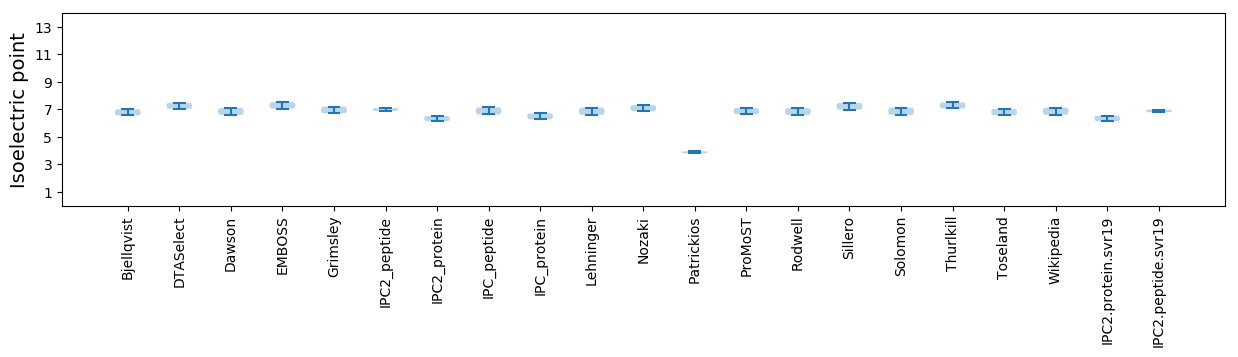
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F8VBM8|F8VBM8_9VIRU Coat protein OS=Pepper cryptic virus 1 OX=369643 PE=4 SV=1
MM1 pKa = 7.88GDD3 pKa = 3.38RR4 pKa = 11.84VNAQDD9 pKa = 4.59DD10 pKa = 4.3DD11 pKa = 4.54TVVPHH16 pKa = 6.01QAPLQPAALQQDD28 pKa = 4.4LTRR31 pKa = 11.84SADD34 pKa = 3.58YY35 pKa = 11.27LLDD38 pKa = 3.36NVRR41 pKa = 11.84IGNHH45 pKa = 4.19RR46 pKa = 11.84QRR48 pKa = 11.84YY49 pKa = 7.93DD50 pKa = 2.67KK51 pKa = 10.73YY52 pKa = 10.4RR53 pKa = 11.84RR54 pKa = 11.84YY55 pKa = 10.27VLLRR59 pKa = 11.84SSEE62 pKa = 4.01IFTSLVAIYY71 pKa = 10.47AHH73 pKa = 6.84IFSSYY78 pKa = 6.8WQHH81 pKa = 5.67FRR83 pKa = 11.84RR84 pKa = 11.84FTDD87 pKa = 3.17QFQAPTGVQLPTFVARR103 pKa = 11.84VYY105 pKa = 10.69ISTWLHH111 pKa = 6.62DD112 pKa = 4.76LYY114 pKa = 11.32CSIRR118 pKa = 11.84EE119 pKa = 4.26ATRR122 pKa = 11.84SISPLAFNEE131 pKa = 4.36RR132 pKa = 11.84YY133 pKa = 9.91SYY135 pKa = 11.27EE136 pKa = 3.94LLPYY140 pKa = 8.73STEE143 pKa = 3.83YY144 pKa = 11.39DD145 pKa = 3.39PFLAFLSMSIKK156 pKa = 8.74PTHH159 pKa = 6.11IQHH162 pKa = 6.04TPEE165 pKa = 3.65NTLWIPILCEE175 pKa = 3.72NYY177 pKa = 10.18DD178 pKa = 3.24WDD180 pKa = 4.94RR181 pKa = 11.84NEE183 pKa = 5.04ANHH186 pKa = 6.33NPFGITNFTLNSNLFYY202 pKa = 11.28GLLAILKK209 pKa = 8.72EE210 pKa = 3.99RR211 pKa = 11.84KK212 pKa = 8.52EE213 pKa = 4.38FKK215 pKa = 10.78LSTLTTNTIGRR226 pKa = 11.84PCWLFDD232 pKa = 3.0WHH234 pKa = 7.77DD235 pKa = 3.96NVQVCAWFPRR245 pKa = 11.84EE246 pKa = 4.33ANFNSQDD253 pKa = 2.95VTAAYY258 pKa = 9.49IIGVACTPKK267 pKa = 10.53LGPSDD272 pKa = 3.96DD273 pKa = 5.02DD274 pKa = 3.06AWKK277 pKa = 10.77YY278 pKa = 9.26YY279 pKa = 11.16ASLNSVPTFTPTEE292 pKa = 3.72PRR294 pKa = 11.84LTNRR298 pKa = 11.84RR299 pKa = 11.84SYY301 pKa = 10.72GAYY304 pKa = 9.19EE305 pKa = 3.77VRR307 pKa = 11.84TRR309 pKa = 11.84EE310 pKa = 3.9TEE312 pKa = 3.83NNYY315 pKa = 9.68FLPDD319 pKa = 3.29SLLNIIEE326 pKa = 5.08DD327 pKa = 4.21FTATGTTQRR336 pKa = 11.84RR337 pKa = 11.84KK338 pKa = 9.5IRR340 pKa = 11.84RR341 pKa = 11.84PSATSASTGAAIIIRR356 pKa = 11.84DD357 pKa = 3.97TPGTASTATTSTTEE371 pKa = 4.15TEE373 pKa = 4.34VTFPPVIRR381 pKa = 11.84TKK383 pKa = 10.39IRR385 pKa = 11.84DD386 pKa = 3.28WYY388 pKa = 7.78YY389 pKa = 8.68HH390 pKa = 5.59SRR392 pKa = 11.84VILEE396 pKa = 4.6LEE398 pKa = 4.5DD399 pKa = 3.79NSRR402 pKa = 11.84TAALRR407 pKa = 11.84MFIIAA412 pKa = 4.64
MM1 pKa = 7.88GDD3 pKa = 3.38RR4 pKa = 11.84VNAQDD9 pKa = 4.59DD10 pKa = 4.3DD11 pKa = 4.54TVVPHH16 pKa = 6.01QAPLQPAALQQDD28 pKa = 4.4LTRR31 pKa = 11.84SADD34 pKa = 3.58YY35 pKa = 11.27LLDD38 pKa = 3.36NVRR41 pKa = 11.84IGNHH45 pKa = 4.19RR46 pKa = 11.84QRR48 pKa = 11.84YY49 pKa = 7.93DD50 pKa = 2.67KK51 pKa = 10.73YY52 pKa = 10.4RR53 pKa = 11.84RR54 pKa = 11.84YY55 pKa = 10.27VLLRR59 pKa = 11.84SSEE62 pKa = 4.01IFTSLVAIYY71 pKa = 10.47AHH73 pKa = 6.84IFSSYY78 pKa = 6.8WQHH81 pKa = 5.67FRR83 pKa = 11.84RR84 pKa = 11.84FTDD87 pKa = 3.17QFQAPTGVQLPTFVARR103 pKa = 11.84VYY105 pKa = 10.69ISTWLHH111 pKa = 6.62DD112 pKa = 4.76LYY114 pKa = 11.32CSIRR118 pKa = 11.84EE119 pKa = 4.26ATRR122 pKa = 11.84SISPLAFNEE131 pKa = 4.36RR132 pKa = 11.84YY133 pKa = 9.91SYY135 pKa = 11.27EE136 pKa = 3.94LLPYY140 pKa = 8.73STEE143 pKa = 3.83YY144 pKa = 11.39DD145 pKa = 3.39PFLAFLSMSIKK156 pKa = 8.74PTHH159 pKa = 6.11IQHH162 pKa = 6.04TPEE165 pKa = 3.65NTLWIPILCEE175 pKa = 3.72NYY177 pKa = 10.18DD178 pKa = 3.24WDD180 pKa = 4.94RR181 pKa = 11.84NEE183 pKa = 5.04ANHH186 pKa = 6.33NPFGITNFTLNSNLFYY202 pKa = 11.28GLLAILKK209 pKa = 8.72EE210 pKa = 3.99RR211 pKa = 11.84KK212 pKa = 8.52EE213 pKa = 4.38FKK215 pKa = 10.78LSTLTTNTIGRR226 pKa = 11.84PCWLFDD232 pKa = 3.0WHH234 pKa = 7.77DD235 pKa = 3.96NVQVCAWFPRR245 pKa = 11.84EE246 pKa = 4.33ANFNSQDD253 pKa = 2.95VTAAYY258 pKa = 9.49IIGVACTPKK267 pKa = 10.53LGPSDD272 pKa = 3.96DD273 pKa = 5.02DD274 pKa = 3.06AWKK277 pKa = 10.77YY278 pKa = 9.26YY279 pKa = 11.16ASLNSVPTFTPTEE292 pKa = 3.72PRR294 pKa = 11.84LTNRR298 pKa = 11.84RR299 pKa = 11.84SYY301 pKa = 10.72GAYY304 pKa = 9.19EE305 pKa = 3.77VRR307 pKa = 11.84TRR309 pKa = 11.84EE310 pKa = 3.9TEE312 pKa = 3.83NNYY315 pKa = 9.68FLPDD319 pKa = 3.29SLLNIIEE326 pKa = 5.08DD327 pKa = 4.21FTATGTTQRR336 pKa = 11.84RR337 pKa = 11.84KK338 pKa = 9.5IRR340 pKa = 11.84RR341 pKa = 11.84PSATSASTGAAIIIRR356 pKa = 11.84DD357 pKa = 3.97TPGTASTATTSTTEE371 pKa = 4.15TEE373 pKa = 4.34VTFPPVIRR381 pKa = 11.84TKK383 pKa = 10.39IRR385 pKa = 11.84DD386 pKa = 3.28WYY388 pKa = 7.78YY389 pKa = 8.68HH390 pKa = 5.59SRR392 pKa = 11.84VILEE396 pKa = 4.6LEE398 pKa = 4.5DD399 pKa = 3.79NSRR402 pKa = 11.84TAALRR407 pKa = 11.84MFIIAA412 pKa = 4.64
Molecular weight: 47.57 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F8VBM8|F8VBM8_9VIRU Coat protein OS=Pepper cryptic virus 1 OX=369643 PE=4 SV=1
MM1 pKa = 7.47VRR3 pKa = 11.84GTLVGYY9 pKa = 10.32DD10 pKa = 3.38YY11 pKa = 11.01TQFQGDD17 pKa = 4.27LVKK20 pKa = 10.09STHH23 pKa = 5.37RR24 pKa = 11.84HH25 pKa = 3.93PHH27 pKa = 4.3VVHH30 pKa = 6.91RR31 pKa = 11.84EE32 pKa = 3.47IATTYY37 pKa = 8.82VDD39 pKa = 3.4QYY41 pKa = 11.03AYY43 pKa = 10.35EE44 pKa = 4.71HH45 pKa = 6.97IEE47 pKa = 4.42TFSSLYY53 pKa = 10.1PEE55 pKa = 6.06LILKK59 pKa = 8.85GWSRR63 pKa = 11.84SYY65 pKa = 10.97YY66 pKa = 10.59LPEE69 pKa = 3.73KK70 pKa = 10.48HH71 pKa = 6.31LAAVLNYY78 pKa = 10.89SMPNVPASQLSQSLYY93 pKa = 9.15RR94 pKa = 11.84QAIEE98 pKa = 3.84SAKK101 pKa = 10.78NGFISLPRR109 pKa = 11.84VKK111 pKa = 10.6AFDD114 pKa = 3.61VLTEE118 pKa = 3.89MDD120 pKa = 3.63QVPFKK125 pKa = 10.65SSSSAGYY132 pKa = 10.35NYY134 pKa = 9.84TGRR137 pKa = 11.84KK138 pKa = 8.81GLIGDD143 pKa = 4.25EE144 pKa = 4.11NHH146 pKa = 6.1SRR148 pKa = 11.84AISIAKK154 pKa = 9.31AVLWSAIKK162 pKa = 10.42DD163 pKa = 3.46DD164 pKa = 4.31GEE166 pKa = 4.7GIEE169 pKa = 4.28HH170 pKa = 7.42VIRR173 pKa = 11.84TSVPDD178 pKa = 3.2VGYY181 pKa = 8.74TRR183 pKa = 11.84TQLTDD188 pKa = 3.73LLEE191 pKa = 4.36KK192 pKa = 10.3TKK194 pKa = 10.27VRR196 pKa = 11.84QVWGRR201 pKa = 11.84AFHH204 pKa = 6.55YY205 pKa = 10.25ILLEE209 pKa = 3.96GLVAYY214 pKa = 8.33PFIQTVMSHH223 pKa = 5.15KK224 pKa = 9.44TFIHH228 pKa = 6.77AGQDD232 pKa = 3.46PLISVPRR239 pKa = 11.84LLSDD243 pKa = 3.79VALNCKK249 pKa = 9.3WIYY252 pKa = 10.66SLDD255 pKa = 3.26WSQFDD260 pKa = 3.45ATVSRR265 pKa = 11.84FEE267 pKa = 3.43IHH269 pKa = 6.71AAFDD273 pKa = 3.98IIKK276 pKa = 10.37SYY278 pKa = 11.8VDD280 pKa = 3.67FPNYY284 pKa = 7.73EE285 pKa = 3.86TEE287 pKa = 3.97QAFEE291 pKa = 3.83ITRR294 pKa = 11.84QLFIHH299 pKa = 6.53KK300 pKa = 9.66KK301 pKa = 8.28VAVPDD306 pKa = 4.33GYY308 pKa = 10.21IYY310 pKa = 10.12EE311 pKa = 4.24SHH313 pKa = 6.77KK314 pKa = 10.8GIPSGSYY321 pKa = 7.44YY322 pKa = 10.2TSLVGSIINYY332 pKa = 9.38LRR334 pKa = 11.84INYY337 pKa = 8.45LWRR340 pKa = 11.84LLTGHH345 pKa = 7.76PPQQCHH351 pKa = 5.2TLGDD355 pKa = 3.97DD356 pKa = 4.19SLVGDD361 pKa = 4.06NSYY364 pKa = 11.54VNPQAIEE371 pKa = 3.66EE372 pKa = 4.17AANKK376 pKa = 9.98LGWHH380 pKa = 6.56FNPDD384 pKa = 2.74KK385 pKa = 9.97TQYY388 pKa = 9.19STVPEE393 pKa = 4.7EE394 pKa = 3.75ITFLGRR400 pKa = 11.84TYY402 pKa = 11.16VGGLNKK408 pKa = 9.58RR409 pKa = 11.84DD410 pKa = 3.34LTKK413 pKa = 10.69CIRR416 pKa = 11.84LLVYY420 pKa = 9.84PEE422 pKa = 4.25YY423 pKa = 9.79PVEE426 pKa = 4.16SGRR429 pKa = 11.84ISAYY433 pKa = 8.84RR434 pKa = 11.84AKK436 pKa = 10.64SIAQDD441 pKa = 3.16AGGLSEE447 pKa = 4.34VLNRR451 pKa = 11.84IADD454 pKa = 3.76KK455 pKa = 10.81LRR457 pKa = 11.84RR458 pKa = 11.84IYY460 pKa = 9.59GTASEE465 pKa = 4.82EE466 pKa = 3.81EE467 pKa = 3.97VPIYY471 pKa = 9.99FKK473 pKa = 10.73RR474 pKa = 11.84YY475 pKa = 8.22VFGVV479 pKa = 3.32
MM1 pKa = 7.47VRR3 pKa = 11.84GTLVGYY9 pKa = 10.32DD10 pKa = 3.38YY11 pKa = 11.01TQFQGDD17 pKa = 4.27LVKK20 pKa = 10.09STHH23 pKa = 5.37RR24 pKa = 11.84HH25 pKa = 3.93PHH27 pKa = 4.3VVHH30 pKa = 6.91RR31 pKa = 11.84EE32 pKa = 3.47IATTYY37 pKa = 8.82VDD39 pKa = 3.4QYY41 pKa = 11.03AYY43 pKa = 10.35EE44 pKa = 4.71HH45 pKa = 6.97IEE47 pKa = 4.42TFSSLYY53 pKa = 10.1PEE55 pKa = 6.06LILKK59 pKa = 8.85GWSRR63 pKa = 11.84SYY65 pKa = 10.97YY66 pKa = 10.59LPEE69 pKa = 3.73KK70 pKa = 10.48HH71 pKa = 6.31LAAVLNYY78 pKa = 10.89SMPNVPASQLSQSLYY93 pKa = 9.15RR94 pKa = 11.84QAIEE98 pKa = 3.84SAKK101 pKa = 10.78NGFISLPRR109 pKa = 11.84VKK111 pKa = 10.6AFDD114 pKa = 3.61VLTEE118 pKa = 3.89MDD120 pKa = 3.63QVPFKK125 pKa = 10.65SSSSAGYY132 pKa = 10.35NYY134 pKa = 9.84TGRR137 pKa = 11.84KK138 pKa = 8.81GLIGDD143 pKa = 4.25EE144 pKa = 4.11NHH146 pKa = 6.1SRR148 pKa = 11.84AISIAKK154 pKa = 9.31AVLWSAIKK162 pKa = 10.42DD163 pKa = 3.46DD164 pKa = 4.31GEE166 pKa = 4.7GIEE169 pKa = 4.28HH170 pKa = 7.42VIRR173 pKa = 11.84TSVPDD178 pKa = 3.2VGYY181 pKa = 8.74TRR183 pKa = 11.84TQLTDD188 pKa = 3.73LLEE191 pKa = 4.36KK192 pKa = 10.3TKK194 pKa = 10.27VRR196 pKa = 11.84QVWGRR201 pKa = 11.84AFHH204 pKa = 6.55YY205 pKa = 10.25ILLEE209 pKa = 3.96GLVAYY214 pKa = 8.33PFIQTVMSHH223 pKa = 5.15KK224 pKa = 9.44TFIHH228 pKa = 6.77AGQDD232 pKa = 3.46PLISVPRR239 pKa = 11.84LLSDD243 pKa = 3.79VALNCKK249 pKa = 9.3WIYY252 pKa = 10.66SLDD255 pKa = 3.26WSQFDD260 pKa = 3.45ATVSRR265 pKa = 11.84FEE267 pKa = 3.43IHH269 pKa = 6.71AAFDD273 pKa = 3.98IIKK276 pKa = 10.37SYY278 pKa = 11.8VDD280 pKa = 3.67FPNYY284 pKa = 7.73EE285 pKa = 3.86TEE287 pKa = 3.97QAFEE291 pKa = 3.83ITRR294 pKa = 11.84QLFIHH299 pKa = 6.53KK300 pKa = 9.66KK301 pKa = 8.28VAVPDD306 pKa = 4.33GYY308 pKa = 10.21IYY310 pKa = 10.12EE311 pKa = 4.24SHH313 pKa = 6.77KK314 pKa = 10.8GIPSGSYY321 pKa = 7.44YY322 pKa = 10.2TSLVGSIINYY332 pKa = 9.38LRR334 pKa = 11.84INYY337 pKa = 8.45LWRR340 pKa = 11.84LLTGHH345 pKa = 7.76PPQQCHH351 pKa = 5.2TLGDD355 pKa = 3.97DD356 pKa = 4.19SLVGDD361 pKa = 4.06NSYY364 pKa = 11.54VNPQAIEE371 pKa = 3.66EE372 pKa = 4.17AANKK376 pKa = 9.98LGWHH380 pKa = 6.56FNPDD384 pKa = 2.74KK385 pKa = 9.97TQYY388 pKa = 9.19STVPEE393 pKa = 4.7EE394 pKa = 3.75ITFLGRR400 pKa = 11.84TYY402 pKa = 11.16VGGLNKK408 pKa = 9.58RR409 pKa = 11.84DD410 pKa = 3.34LTKK413 pKa = 10.69CIRR416 pKa = 11.84LLVYY420 pKa = 9.84PEE422 pKa = 4.25YY423 pKa = 9.79PVEE426 pKa = 4.16SGRR429 pKa = 11.84ISAYY433 pKa = 8.84RR434 pKa = 11.84AKK436 pKa = 10.64SIAQDD441 pKa = 3.16AGGLSEE447 pKa = 4.34VLNRR451 pKa = 11.84IADD454 pKa = 3.76KK455 pKa = 10.81LRR457 pKa = 11.84RR458 pKa = 11.84IYY460 pKa = 9.59GTASEE465 pKa = 4.82EE466 pKa = 3.81EE467 pKa = 3.97VPIYY471 pKa = 9.99FKK473 pKa = 10.73RR474 pKa = 11.84YY475 pKa = 8.22VFGVV479 pKa = 3.32
Molecular weight: 54.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
891 |
412 |
479 |
445.5 |
51.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.071 ± 0.441 | 0.898 ± 0.2 |
5.387 ± 0.277 | 5.275 ± 0.266 |
4.377 ± 0.456 | 4.826 ± 1.212 |
3.03 ± 0.382 | 6.846 ± 0.185 |
3.704 ± 0.962 | 8.642 ± 0.093 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
0.786 ± 0.036 | 4.153 ± 0.752 |
5.163 ± 0.42 | 3.704 ± 0.194 |
6.622 ± 0.879 | 7.632 ± 0.376 |
7.856 ± 1.634 | 6.061 ± 1.071 |
1.796 ± 0.246 | 6.173 ± 0.528 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
