
Geodermatophilus sp. Leaf369
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Geodermatophilales; Geodermatophilaceae; Geodermatophilus; unclassified Geodermatophilus
Average proteome isoelectric point is 5.96
Get precalculated fractions of proteins
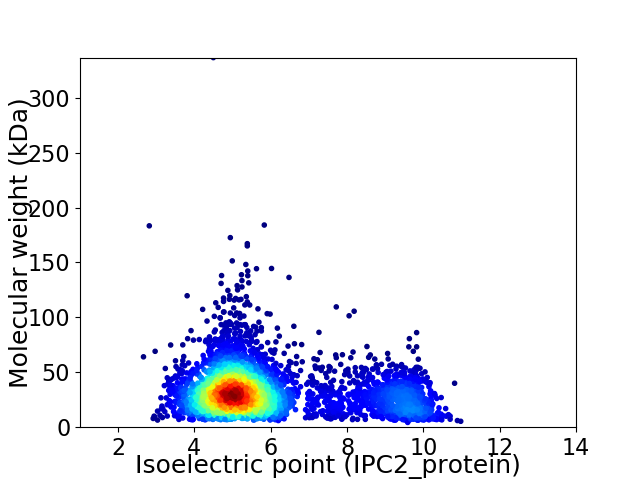
Virtual 2D-PAGE plot for 4093 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
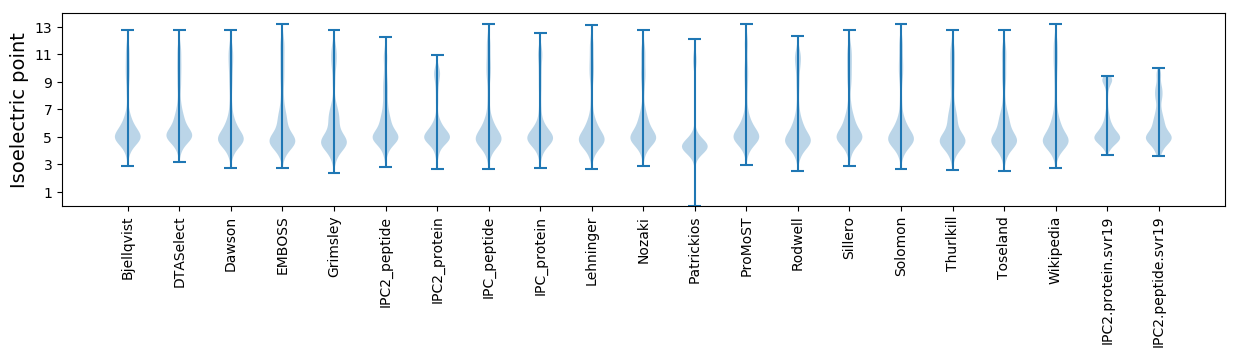
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q5VF77|A0A0Q5VF77_9ACTN Diacylglycerol kinase OS=Geodermatophilus sp. Leaf369 OX=1736354 GN=ASG36_00055 PE=4 SV=1
MM1 pKa = 7.4AVAGGLVLLAGCTGGSSSSGQADD24 pKa = 3.9GSSAPTSEE32 pKa = 4.88APAAQLAVSPTDD44 pKa = 3.28GTADD48 pKa = 3.49VSPTTPVQLAVTDD61 pKa = 4.2GAIGEE66 pKa = 4.46VTVTDD71 pKa = 3.45AAGTVVPGAVAEE83 pKa = 4.55APADD87 pKa = 3.55PAAPTTAAPGATTSVWTPTDD107 pKa = 3.37PLAYY111 pKa = 8.12GTTYY115 pKa = 10.48TVDD118 pKa = 3.24ATATNADD125 pKa = 4.06DD126 pKa = 5.1AEE128 pKa = 4.55TDD130 pKa = 3.06ASTTFSTVTPASVTTPSIGPLDD152 pKa = 3.94GTTVGVGMPIRR163 pKa = 11.84VFFDD167 pKa = 3.43QPVTDD172 pKa = 3.76QAAVEE177 pKa = 4.19SHH179 pKa = 6.75LLVTSSTPTDD189 pKa = 3.87GRR191 pKa = 11.84WNWVSSSEE199 pKa = 4.03VHH201 pKa = 6.17FRR203 pKa = 11.84PSTYY207 pKa = 9.38WPANTTVTLDD217 pKa = 3.22ADD219 pKa = 4.4LYY221 pKa = 11.56GVDD224 pKa = 4.09TGDD227 pKa = 4.72GIWGEE232 pKa = 4.09KK233 pKa = 10.15DD234 pKa = 2.83RR235 pKa = 11.84TVTFTVGEE243 pKa = 4.19KK244 pKa = 10.09HH245 pKa = 6.7VSVADD250 pKa = 4.05AGSHH254 pKa = 5.1TLTVYY259 pKa = 10.97DD260 pKa = 3.94SDD262 pKa = 4.34QVVQTYY268 pKa = 9.98PMSAGSSEE276 pKa = 3.74NPTRR280 pKa = 11.84NGAHH284 pKa = 5.48VVLEE288 pKa = 4.25KK289 pKa = 11.45YY290 pKa = 10.56EE291 pKa = 4.97DD292 pKa = 3.26ITMDD296 pKa = 2.88SSTFGLAVDD305 pKa = 3.73AAGGYY310 pKa = 7.88RR311 pKa = 11.84TDD313 pKa = 2.79VQYY316 pKa = 10.3ATRR319 pKa = 11.84ISNNGEE325 pKa = 3.95FVHH328 pKa = 6.5AAPWSVGQQGSSNVSHH344 pKa = 6.66GCINLSTDD352 pKa = 3.25RR353 pKa = 11.84AQWFYY358 pKa = 11.71DD359 pKa = 4.28FSQPGDD365 pKa = 3.43VVEE368 pKa = 4.59VVNSIGPTLSPVDD381 pKa = 3.41GDD383 pKa = 3.57IYY385 pKa = 11.3DD386 pKa = 3.74WAIPWDD392 pKa = 3.49QWVAGSALSS401 pKa = 3.48
MM1 pKa = 7.4AVAGGLVLLAGCTGGSSSSGQADD24 pKa = 3.9GSSAPTSEE32 pKa = 4.88APAAQLAVSPTDD44 pKa = 3.28GTADD48 pKa = 3.49VSPTTPVQLAVTDD61 pKa = 4.2GAIGEE66 pKa = 4.46VTVTDD71 pKa = 3.45AAGTVVPGAVAEE83 pKa = 4.55APADD87 pKa = 3.55PAAPTTAAPGATTSVWTPTDD107 pKa = 3.37PLAYY111 pKa = 8.12GTTYY115 pKa = 10.48TVDD118 pKa = 3.24ATATNADD125 pKa = 4.06DD126 pKa = 5.1AEE128 pKa = 4.55TDD130 pKa = 3.06ASTTFSTVTPASVTTPSIGPLDD152 pKa = 3.94GTTVGVGMPIRR163 pKa = 11.84VFFDD167 pKa = 3.43QPVTDD172 pKa = 3.76QAAVEE177 pKa = 4.19SHH179 pKa = 6.75LLVTSSTPTDD189 pKa = 3.87GRR191 pKa = 11.84WNWVSSSEE199 pKa = 4.03VHH201 pKa = 6.17FRR203 pKa = 11.84PSTYY207 pKa = 9.38WPANTTVTLDD217 pKa = 3.22ADD219 pKa = 4.4LYY221 pKa = 11.56GVDD224 pKa = 4.09TGDD227 pKa = 4.72GIWGEE232 pKa = 4.09KK233 pKa = 10.15DD234 pKa = 2.83RR235 pKa = 11.84TVTFTVGEE243 pKa = 4.19KK244 pKa = 10.09HH245 pKa = 6.7VSVADD250 pKa = 4.05AGSHH254 pKa = 5.1TLTVYY259 pKa = 10.97DD260 pKa = 3.94SDD262 pKa = 4.34QVVQTYY268 pKa = 9.98PMSAGSSEE276 pKa = 3.74NPTRR280 pKa = 11.84NGAHH284 pKa = 5.48VVLEE288 pKa = 4.25KK289 pKa = 11.45YY290 pKa = 10.56EE291 pKa = 4.97DD292 pKa = 3.26ITMDD296 pKa = 2.88SSTFGLAVDD305 pKa = 3.73AAGGYY310 pKa = 7.88RR311 pKa = 11.84TDD313 pKa = 2.79VQYY316 pKa = 10.3ATRR319 pKa = 11.84ISNNGEE325 pKa = 3.95FVHH328 pKa = 6.5AAPWSVGQQGSSNVSHH344 pKa = 6.66GCINLSTDD352 pKa = 3.25RR353 pKa = 11.84AQWFYY358 pKa = 11.71DD359 pKa = 4.28FSQPGDD365 pKa = 3.43VVEE368 pKa = 4.59VVNSIGPTLSPVDD381 pKa = 3.41GDD383 pKa = 3.57IYY385 pKa = 11.3DD386 pKa = 3.74WAIPWDD392 pKa = 3.49QWVAGSALSS401 pKa = 3.48
Molecular weight: 41.18 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q5VAK0|A0A0Q5VAK0_9ACTN Uncharacterized protein OS=Geodermatophilus sp. Leaf369 OX=1736354 GN=ASG36_09915 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84SKK15 pKa = 8.58THH17 pKa = 5.46GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILAGRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 10.08GRR40 pKa = 11.84EE41 pKa = 3.61KK42 pKa = 10.93LSAA45 pKa = 3.78
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84SKK15 pKa = 8.58THH17 pKa = 5.46GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILAGRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 10.08GRR40 pKa = 11.84EE41 pKa = 3.61KK42 pKa = 10.93LSAA45 pKa = 3.78
Molecular weight: 5.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1301898 |
37 |
3380 |
318.1 |
33.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.309 ± 0.061 | 0.642 ± 0.01 |
6.452 ± 0.031 | 5.263 ± 0.038 |
2.539 ± 0.023 | 9.726 ± 0.036 |
2.034 ± 0.019 | 2.538 ± 0.026 |
1.364 ± 0.026 | 10.719 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.527 ± 0.014 | 1.376 ± 0.018 |
6.033 ± 0.038 | 2.803 ± 0.02 |
7.577 ± 0.04 | 4.963 ± 0.026 |
6.593 ± 0.04 | 10.425 ± 0.037 |
1.468 ± 0.015 | 1.648 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
