
Halospina denitrificans
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Gammaproteobacteria incertae sedis; Halospina
Average proteome isoelectric point is 5.79
Get precalculated fractions of proteins
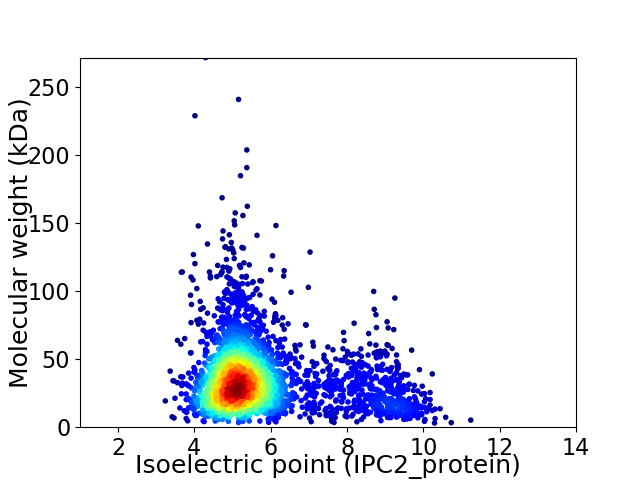
Virtual 2D-PAGE plot for 3014 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4V3ERE7|A0A4V3ERE7_9GAMM Cobyric acid synthase OS=Halospina denitrificans OX=332522 GN=cobQ PE=3 SV=1
MM1 pKa = 7.34NRR3 pKa = 11.84NLLAAAVAASVTAAAPSLAVASTNISAGVWASYY36 pKa = 10.6RR37 pKa = 11.84YY38 pKa = 8.54VTDD41 pKa = 3.25SDD43 pKa = 3.89FAAPAFNSDD52 pKa = 3.51LDD54 pKa = 4.01EE55 pKa = 4.45EE56 pKa = 4.72TGGDD60 pKa = 3.22IGRR63 pKa = 11.84EE64 pKa = 3.95SLILYY69 pKa = 9.6GDD71 pKa = 3.72HH72 pKa = 5.78TQEE75 pKa = 4.46DD76 pKa = 5.46GPWRR80 pKa = 11.84LSAEE84 pKa = 4.1VRR86 pKa = 11.84FGPGSFSDD94 pKa = 4.01PDD96 pKa = 3.81NNSTGSFTAVHH107 pKa = 6.39KK108 pKa = 10.38LWAGRR113 pKa = 11.84SIGEE117 pKa = 3.99NWDD120 pKa = 3.38MKK122 pKa = 10.12IGKK125 pKa = 9.27SEE127 pKa = 4.27VPFGWKK133 pKa = 7.79TVNFWPGDD141 pKa = 3.84LLLGGYY147 pKa = 9.34GDD149 pKa = 4.21QMDD152 pKa = 4.07VGAKK156 pKa = 7.99FTGDD160 pKa = 3.07VGGWDD165 pKa = 3.28LAAAYY170 pKa = 8.07YY171 pKa = 10.1HH172 pKa = 6.05QDD174 pKa = 3.14DD175 pKa = 3.96WGEE178 pKa = 4.13TSTDD182 pKa = 3.58TIDD185 pKa = 5.9DD186 pKa = 3.87NGHH189 pKa = 6.22WGSSTTYY196 pKa = 10.48RR197 pKa = 11.84KK198 pKa = 9.48IQTGVADD205 pKa = 4.12ASYY208 pKa = 10.12TFGSDD213 pKa = 2.88GGSSAFGLSAQAGKK227 pKa = 9.7LQDD230 pKa = 3.89LTDD233 pKa = 3.89HH234 pKa = 7.04ANACTTSCTDD244 pKa = 4.92LDD246 pKa = 4.61DD247 pKa = 5.1GSHH250 pKa = 6.38NALALYY256 pKa = 10.65YY257 pKa = 9.39EE258 pKa = 4.8GAYY261 pKa = 10.6GPFTAKK267 pKa = 10.65AEE269 pKa = 3.98AMQVNRR275 pKa = 11.84DD276 pKa = 3.66VPNVANDD283 pKa = 3.55IEE285 pKa = 4.3NTRR288 pKa = 11.84YY289 pKa = 10.2ALQGGYY295 pKa = 10.0SVGNWFMFLDD305 pKa = 3.97ATAANTDD312 pKa = 3.24TDD314 pKa = 4.18GNQADD319 pKa = 3.95TVYY322 pKa = 10.77AFSPGVTYY330 pKa = 10.29DD331 pKa = 3.89YY332 pKa = 11.38GPGWIYY338 pKa = 11.09LEE340 pKa = 4.05YY341 pKa = 10.5LKK343 pKa = 10.94SDD345 pKa = 3.35GDD347 pKa = 3.37IDD349 pKa = 4.66ANGDD353 pKa = 3.09IYY355 pKa = 11.14EE356 pKa = 4.44ADD358 pKa = 3.62FDD360 pKa = 4.67AMYY363 pKa = 9.89VTMDD367 pKa = 3.3YY368 pKa = 11.39YY369 pKa = 11.23FF370 pKa = 4.84
MM1 pKa = 7.34NRR3 pKa = 11.84NLLAAAVAASVTAAAPSLAVASTNISAGVWASYY36 pKa = 10.6RR37 pKa = 11.84YY38 pKa = 8.54VTDD41 pKa = 3.25SDD43 pKa = 3.89FAAPAFNSDD52 pKa = 3.51LDD54 pKa = 4.01EE55 pKa = 4.45EE56 pKa = 4.72TGGDD60 pKa = 3.22IGRR63 pKa = 11.84EE64 pKa = 3.95SLILYY69 pKa = 9.6GDD71 pKa = 3.72HH72 pKa = 5.78TQEE75 pKa = 4.46DD76 pKa = 5.46GPWRR80 pKa = 11.84LSAEE84 pKa = 4.1VRR86 pKa = 11.84FGPGSFSDD94 pKa = 4.01PDD96 pKa = 3.81NNSTGSFTAVHH107 pKa = 6.39KK108 pKa = 10.38LWAGRR113 pKa = 11.84SIGEE117 pKa = 3.99NWDD120 pKa = 3.38MKK122 pKa = 10.12IGKK125 pKa = 9.27SEE127 pKa = 4.27VPFGWKK133 pKa = 7.79TVNFWPGDD141 pKa = 3.84LLLGGYY147 pKa = 9.34GDD149 pKa = 4.21QMDD152 pKa = 4.07VGAKK156 pKa = 7.99FTGDD160 pKa = 3.07VGGWDD165 pKa = 3.28LAAAYY170 pKa = 8.07YY171 pKa = 10.1HH172 pKa = 6.05QDD174 pKa = 3.14DD175 pKa = 3.96WGEE178 pKa = 4.13TSTDD182 pKa = 3.58TIDD185 pKa = 5.9DD186 pKa = 3.87NGHH189 pKa = 6.22WGSSTTYY196 pKa = 10.48RR197 pKa = 11.84KK198 pKa = 9.48IQTGVADD205 pKa = 4.12ASYY208 pKa = 10.12TFGSDD213 pKa = 2.88GGSSAFGLSAQAGKK227 pKa = 9.7LQDD230 pKa = 3.89LTDD233 pKa = 3.89HH234 pKa = 7.04ANACTTSCTDD244 pKa = 4.92LDD246 pKa = 4.61DD247 pKa = 5.1GSHH250 pKa = 6.38NALALYY256 pKa = 10.65YY257 pKa = 9.39EE258 pKa = 4.8GAYY261 pKa = 10.6GPFTAKK267 pKa = 10.65AEE269 pKa = 3.98AMQVNRR275 pKa = 11.84DD276 pKa = 3.66VPNVANDD283 pKa = 3.55IEE285 pKa = 4.3NTRR288 pKa = 11.84YY289 pKa = 10.2ALQGGYY295 pKa = 10.0SVGNWFMFLDD305 pKa = 3.97ATAANTDD312 pKa = 3.24TDD314 pKa = 4.18GNQADD319 pKa = 3.95TVYY322 pKa = 10.77AFSPGVTYY330 pKa = 10.29DD331 pKa = 3.89YY332 pKa = 11.38GPGWIYY338 pKa = 11.09LEE340 pKa = 4.05YY341 pKa = 10.5LKK343 pKa = 10.94SDD345 pKa = 3.35GDD347 pKa = 3.37IDD349 pKa = 4.66ANGDD353 pKa = 3.09IYY355 pKa = 11.14EE356 pKa = 4.44ADD358 pKa = 3.62FDD360 pKa = 4.67AMYY363 pKa = 9.89VTMDD367 pKa = 3.3YY368 pKa = 11.39YY369 pKa = 11.23FF370 pKa = 4.84
Molecular weight: 39.79 kDa
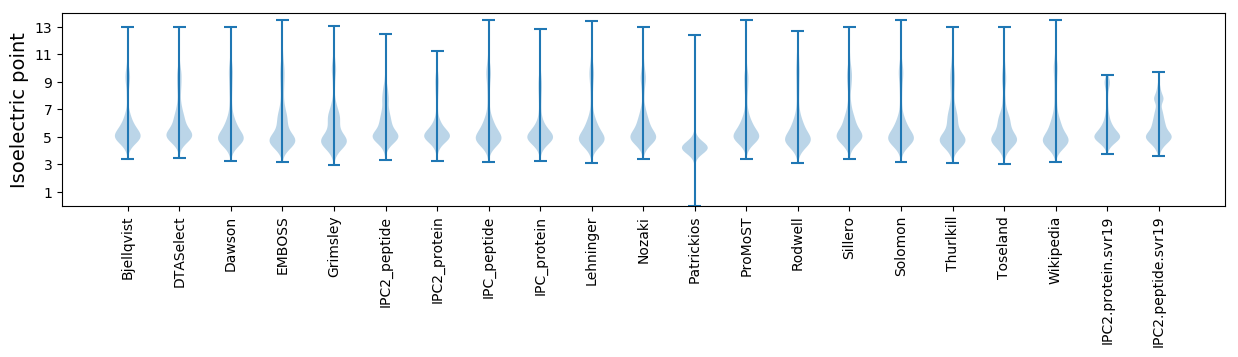
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4R7K171|A0A4R7K171_9GAMM CBS domain-containing protein OS=Halospina denitrificans OX=332522 GN=DES49_0286 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.42RR12 pKa = 11.84KK13 pKa = 9.04RR14 pKa = 11.84AHH16 pKa = 6.12GFRR19 pKa = 11.84ARR21 pKa = 11.84MATQNGRR28 pKa = 11.84QVIARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.73GRR39 pKa = 11.84KK40 pKa = 8.55RR41 pKa = 11.84LSVV44 pKa = 3.2
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.42RR12 pKa = 11.84KK13 pKa = 9.04RR14 pKa = 11.84AHH16 pKa = 6.12GFRR19 pKa = 11.84ARR21 pKa = 11.84MATQNGRR28 pKa = 11.84QVIARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.73GRR39 pKa = 11.84KK40 pKa = 8.55RR41 pKa = 11.84LSVV44 pKa = 3.2
Molecular weight: 5.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1010808 |
29 |
2461 |
335.4 |
37.14 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.576 ± 0.049 | 0.906 ± 0.015 |
6.058 ± 0.038 | 7.307 ± 0.049 |
3.619 ± 0.029 | 8.047 ± 0.039 |
2.366 ± 0.024 | 4.909 ± 0.029 |
2.994 ± 0.036 | 10.523 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.563 ± 0.02 | 3.14 ± 0.022 |
4.85 ± 0.031 | 3.957 ± 0.028 |
7.19 ± 0.042 | 5.803 ± 0.031 |
5.163 ± 0.033 | 7.006 ± 0.039 |
1.427 ± 0.019 | 2.596 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
