
Periplaneta fuliginosa densovirus (isolate Guo/2000) (PfDNV)
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Quintoviricetes; Piccovirales; Parvoviridae; Densovirinae; Pefuambidensovirus; Blattodean pefuambidensovirus 1
Average proteome isoelectric point is 5.44
Get precalculated fractions of proteins
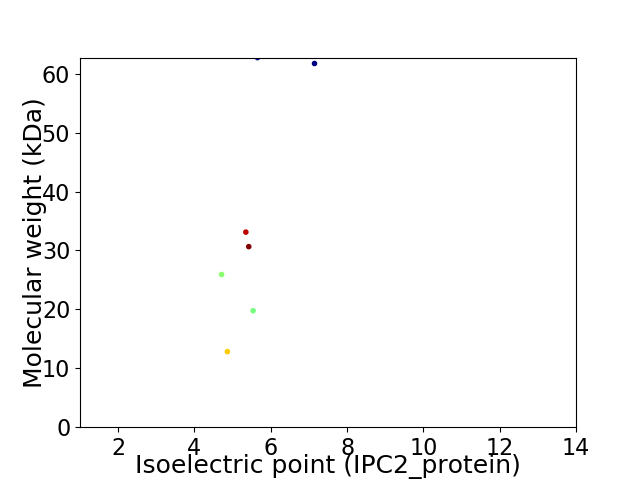
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q9QCZ5|Q9QCZ5_PFDNG Structural protein OS=Periplaneta fuliginosa densovirus (isolate Guo/2000) OX=648333 PE=4 SV=1
MM1 pKa = 7.33GSEE4 pKa = 4.12LRR6 pKa = 11.84PLISEE11 pKa = 4.26VQQEE15 pKa = 4.36RR16 pKa = 11.84FSTDD20 pKa = 2.34TNRR23 pKa = 11.84IYY25 pKa = 10.87NAGYY29 pKa = 10.11SEE31 pKa = 3.68IRR33 pKa = 11.84EE34 pKa = 4.4PNWGLRR40 pKa = 11.84NRR42 pKa = 11.84ISGWSEE48 pKa = 3.38RR49 pKa = 11.84VRR51 pKa = 11.84EE52 pKa = 3.99LTRR55 pKa = 11.84PNNYY59 pKa = 9.96SVIEE63 pKa = 4.31GSDD66 pKa = 3.08IEE68 pKa = 4.97LPISVDD74 pKa = 3.72DD75 pKa = 4.74DD76 pKa = 3.65VHH78 pKa = 8.34IPIEE82 pKa = 4.36EE83 pKa = 4.05EE84 pKa = 3.95TSFSVSVPEE93 pKa = 4.02ATEE96 pKa = 4.13TTGLLGTSVISAGTGVAGGAATGLAASDD124 pKa = 3.34IGLGAGAVIGSGIVGYY140 pKa = 9.86LGHH143 pKa = 7.1KK144 pKa = 9.44IISGLTYY151 pKa = 10.18PFHH154 pKa = 7.51HH155 pKa = 6.69YY156 pKa = 10.32LGPGNPLDD164 pKa = 3.91NNEE167 pKa = 4.12PVDD170 pKa = 4.24RR171 pKa = 11.84DD172 pKa = 3.51DD173 pKa = 6.49AIAEE177 pKa = 4.24EE178 pKa = 4.41HH179 pKa = 6.91DD180 pKa = 3.53KK181 pKa = 11.31AYY183 pKa = 10.83ANAKK187 pKa = 10.21SSIDD191 pKa = 4.1VINADD196 pKa = 4.27KK197 pKa = 10.93KK198 pKa = 11.08AIDD201 pKa = 3.85HH202 pKa = 6.39FSEE205 pKa = 5.07DD206 pKa = 3.6FEE208 pKa = 5.85KK209 pKa = 11.15NGNLHH214 pKa = 6.05SLIGKK219 pKa = 8.14TGLQIKK225 pKa = 7.1TAIEE229 pKa = 3.63QRR231 pKa = 11.84FGVIYY236 pKa = 9.67PSVSGTT242 pKa = 3.25
MM1 pKa = 7.33GSEE4 pKa = 4.12LRR6 pKa = 11.84PLISEE11 pKa = 4.26VQQEE15 pKa = 4.36RR16 pKa = 11.84FSTDD20 pKa = 2.34TNRR23 pKa = 11.84IYY25 pKa = 10.87NAGYY29 pKa = 10.11SEE31 pKa = 3.68IRR33 pKa = 11.84EE34 pKa = 4.4PNWGLRR40 pKa = 11.84NRR42 pKa = 11.84ISGWSEE48 pKa = 3.38RR49 pKa = 11.84VRR51 pKa = 11.84EE52 pKa = 3.99LTRR55 pKa = 11.84PNNYY59 pKa = 9.96SVIEE63 pKa = 4.31GSDD66 pKa = 3.08IEE68 pKa = 4.97LPISVDD74 pKa = 3.72DD75 pKa = 4.74DD76 pKa = 3.65VHH78 pKa = 8.34IPIEE82 pKa = 4.36EE83 pKa = 4.05EE84 pKa = 3.95TSFSVSVPEE93 pKa = 4.02ATEE96 pKa = 4.13TTGLLGTSVISAGTGVAGGAATGLAASDD124 pKa = 3.34IGLGAGAVIGSGIVGYY140 pKa = 9.86LGHH143 pKa = 7.1KK144 pKa = 9.44IISGLTYY151 pKa = 10.18PFHH154 pKa = 7.51HH155 pKa = 6.69YY156 pKa = 10.32LGPGNPLDD164 pKa = 3.91NNEE167 pKa = 4.12PVDD170 pKa = 4.24RR171 pKa = 11.84DD172 pKa = 3.51DD173 pKa = 6.49AIAEE177 pKa = 4.24EE178 pKa = 4.41HH179 pKa = 6.91DD180 pKa = 3.53KK181 pKa = 11.31AYY183 pKa = 10.83ANAKK187 pKa = 10.21SSIDD191 pKa = 4.1VINADD196 pKa = 4.27KK197 pKa = 10.93KK198 pKa = 11.08AIDD201 pKa = 3.85HH202 pKa = 6.39FSEE205 pKa = 5.07DD206 pKa = 3.6FEE208 pKa = 5.85KK209 pKa = 11.15NGNLHH214 pKa = 6.05SLIGKK219 pKa = 8.14TGLQIKK225 pKa = 7.1TAIEE229 pKa = 3.63QRR231 pKa = 11.84FGVIYY236 pKa = 9.67PSVSGTT242 pKa = 3.25
Molecular weight: 25.92 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q77NU9|Q77NU9_PFDNG Uncharacterized protein OS=Periplaneta fuliginosa densovirus (isolate Guo/2000) OX=648333 PE=4 SV=1
MM1 pKa = 7.54QDD3 pKa = 3.55GKK5 pKa = 10.85QPDD8 pKa = 3.41NSNYY12 pKa = 9.52VPALDD17 pKa = 3.99DD18 pKa = 3.31TMGTVWQRR26 pKa = 11.84MRR28 pKa = 11.84DD29 pKa = 3.51DD30 pKa = 5.09AGFLDD35 pKa = 3.92EE36 pKa = 4.31TARR39 pKa = 11.84DD40 pKa = 3.68GFGGFFSRR48 pKa = 11.84GVEE51 pKa = 3.9NSVSSDD57 pKa = 3.59GEE59 pKa = 3.82ILGPQQQLQRR69 pKa = 11.84DD70 pKa = 3.53IVSRR74 pKa = 11.84IKK76 pKa = 9.89RR77 pKa = 11.84TPFGPFRR84 pKa = 11.84RR85 pKa = 11.84YY86 pKa = 10.32LSDD89 pKa = 4.25VLACDD94 pKa = 4.73DD95 pKa = 3.67SRR97 pKa = 11.84EE98 pKa = 4.19ANRR101 pKa = 11.84IARR104 pKa = 11.84QVIRR108 pKa = 11.84TARR111 pKa = 11.84DD112 pKa = 3.41YY113 pKa = 11.22PGNLCLISTHH123 pKa = 7.15DD124 pKa = 3.66DD125 pKa = 3.48HH126 pKa = 8.86VHH128 pKa = 5.24VVHH131 pKa = 7.37DD132 pKa = 4.11CAYY135 pKa = 10.46SDD137 pKa = 4.05GSCRR141 pKa = 11.84CKK143 pKa = 10.0ILKK146 pKa = 10.32EE147 pKa = 3.97EE148 pKa = 4.06TVKK151 pKa = 10.91AKK153 pKa = 10.44RR154 pKa = 11.84RR155 pKa = 11.84GAIRR159 pKa = 11.84KK160 pKa = 8.64RR161 pKa = 11.84KK162 pKa = 9.17AISSIASDD170 pKa = 3.1QWDD173 pKa = 4.35DD174 pKa = 3.66IILYY178 pKa = 9.02FISNGRR184 pKa = 11.84WLQFAKK190 pKa = 10.77VGGSVVGLPYY200 pKa = 10.41GYY202 pKa = 9.95QALQEE207 pKa = 4.55RR208 pKa = 11.84GHH210 pKa = 6.05QGQDD214 pKa = 2.45SGQILGTCTSTGSSEE229 pKa = 4.27LCGEE233 pKa = 4.2SSIEE237 pKa = 4.03EE238 pKa = 4.17VTGKK242 pKa = 9.27SARR245 pKa = 11.84INSRR249 pKa = 11.84DD250 pKa = 2.75GRR252 pKa = 11.84RR253 pKa = 11.84RR254 pKa = 11.84RR255 pKa = 11.84RR256 pKa = 11.84RR257 pKa = 11.84ASDD260 pKa = 2.69IRR262 pKa = 11.84EE263 pKa = 3.93EE264 pKa = 3.97MEE266 pKa = 4.8KK267 pKa = 10.06IVNEE271 pKa = 4.07NPVCPLAGIISTRR284 pKa = 11.84QWLTHH289 pKa = 6.02KK290 pKa = 10.49DD291 pKa = 3.53LRR293 pKa = 11.84YY294 pKa = 10.07LRR296 pKa = 11.84ADD298 pKa = 3.39NEE300 pKa = 4.81TVKK303 pKa = 10.98SFLDD307 pKa = 3.55AKK309 pKa = 10.51SEE311 pKa = 4.0EE312 pKa = 3.92MCYY315 pKa = 7.77WTLHH319 pKa = 6.43DD320 pKa = 5.31FNTFYY325 pKa = 10.95SQPYY329 pKa = 9.1CNPTFMAGYY338 pKa = 9.8QPLEE342 pKa = 4.06DD343 pKa = 4.26KK344 pKa = 10.85YY345 pKa = 11.68YY346 pKa = 10.98SVNDD350 pKa = 3.56SLTILNKK357 pKa = 10.31LLAYY361 pKa = 10.42QFDD364 pKa = 4.54DD365 pKa = 4.23DD366 pKa = 4.33VVQIKK371 pKa = 10.75SFLNTFYY378 pKa = 11.37NVLEE382 pKa = 4.39RR383 pKa = 11.84KK384 pKa = 9.35LPKK387 pKa = 10.55CNTICVWSPPSAGKK401 pKa = 10.0NFFFDD406 pKa = 3.69VYY408 pKa = 10.89LHH410 pKa = 6.09YY411 pKa = 11.21LMNYY415 pKa = 8.03GQLGIMNKK423 pKa = 8.69TNNFSLQEE431 pKa = 4.05ATSKK435 pKa = 10.72RR436 pKa = 11.84VLLWNEE442 pKa = 3.63PNYY445 pKa = 10.81EE446 pKa = 3.97DD447 pKa = 6.12AYY449 pKa = 10.07TDD451 pKa = 3.63TLKK454 pKa = 10.55MLTGGDD460 pKa = 3.89ALCVRR465 pKa = 11.84VKK467 pKa = 10.36QKK469 pKa = 10.85KK470 pKa = 9.01DD471 pKa = 2.96CHH473 pKa = 6.43VYY475 pKa = 8.51KK476 pKa = 10.51TPLIVLTNNMIGFMHH491 pKa = 7.01EE492 pKa = 3.82LAFVDD497 pKa = 3.87RR498 pKa = 11.84VKK500 pKa = 10.84VYY502 pKa = 9.69RR503 pKa = 11.84WKK505 pKa = 10.37QAPFLAEE512 pKa = 4.06YY513 pKa = 10.07NKK515 pKa = 10.57KK516 pKa = 10.12PNPLVAFEE524 pKa = 4.39ILVYY528 pKa = 10.11WEE530 pKa = 4.46IIPRR534 pKa = 11.84IIEE537 pKa = 3.79QQ538 pKa = 3.25
MM1 pKa = 7.54QDD3 pKa = 3.55GKK5 pKa = 10.85QPDD8 pKa = 3.41NSNYY12 pKa = 9.52VPALDD17 pKa = 3.99DD18 pKa = 3.31TMGTVWQRR26 pKa = 11.84MRR28 pKa = 11.84DD29 pKa = 3.51DD30 pKa = 5.09AGFLDD35 pKa = 3.92EE36 pKa = 4.31TARR39 pKa = 11.84DD40 pKa = 3.68GFGGFFSRR48 pKa = 11.84GVEE51 pKa = 3.9NSVSSDD57 pKa = 3.59GEE59 pKa = 3.82ILGPQQQLQRR69 pKa = 11.84DD70 pKa = 3.53IVSRR74 pKa = 11.84IKK76 pKa = 9.89RR77 pKa = 11.84TPFGPFRR84 pKa = 11.84RR85 pKa = 11.84YY86 pKa = 10.32LSDD89 pKa = 4.25VLACDD94 pKa = 4.73DD95 pKa = 3.67SRR97 pKa = 11.84EE98 pKa = 4.19ANRR101 pKa = 11.84IARR104 pKa = 11.84QVIRR108 pKa = 11.84TARR111 pKa = 11.84DD112 pKa = 3.41YY113 pKa = 11.22PGNLCLISTHH123 pKa = 7.15DD124 pKa = 3.66DD125 pKa = 3.48HH126 pKa = 8.86VHH128 pKa = 5.24VVHH131 pKa = 7.37DD132 pKa = 4.11CAYY135 pKa = 10.46SDD137 pKa = 4.05GSCRR141 pKa = 11.84CKK143 pKa = 10.0ILKK146 pKa = 10.32EE147 pKa = 3.97EE148 pKa = 4.06TVKK151 pKa = 10.91AKK153 pKa = 10.44RR154 pKa = 11.84RR155 pKa = 11.84GAIRR159 pKa = 11.84KK160 pKa = 8.64RR161 pKa = 11.84KK162 pKa = 9.17AISSIASDD170 pKa = 3.1QWDD173 pKa = 4.35DD174 pKa = 3.66IILYY178 pKa = 9.02FISNGRR184 pKa = 11.84WLQFAKK190 pKa = 10.77VGGSVVGLPYY200 pKa = 10.41GYY202 pKa = 9.95QALQEE207 pKa = 4.55RR208 pKa = 11.84GHH210 pKa = 6.05QGQDD214 pKa = 2.45SGQILGTCTSTGSSEE229 pKa = 4.27LCGEE233 pKa = 4.2SSIEE237 pKa = 4.03EE238 pKa = 4.17VTGKK242 pKa = 9.27SARR245 pKa = 11.84INSRR249 pKa = 11.84DD250 pKa = 2.75GRR252 pKa = 11.84RR253 pKa = 11.84RR254 pKa = 11.84RR255 pKa = 11.84RR256 pKa = 11.84RR257 pKa = 11.84ASDD260 pKa = 2.69IRR262 pKa = 11.84EE263 pKa = 3.93EE264 pKa = 3.97MEE266 pKa = 4.8KK267 pKa = 10.06IVNEE271 pKa = 4.07NPVCPLAGIISTRR284 pKa = 11.84QWLTHH289 pKa = 6.02KK290 pKa = 10.49DD291 pKa = 3.53LRR293 pKa = 11.84YY294 pKa = 10.07LRR296 pKa = 11.84ADD298 pKa = 3.39NEE300 pKa = 4.81TVKK303 pKa = 10.98SFLDD307 pKa = 3.55AKK309 pKa = 10.51SEE311 pKa = 4.0EE312 pKa = 3.92MCYY315 pKa = 7.77WTLHH319 pKa = 6.43DD320 pKa = 5.31FNTFYY325 pKa = 10.95SQPYY329 pKa = 9.1CNPTFMAGYY338 pKa = 9.8QPLEE342 pKa = 4.06DD343 pKa = 4.26KK344 pKa = 10.85YY345 pKa = 11.68YY346 pKa = 10.98SVNDD350 pKa = 3.56SLTILNKK357 pKa = 10.31LLAYY361 pKa = 10.42QFDD364 pKa = 4.54DD365 pKa = 4.23DD366 pKa = 4.33VVQIKK371 pKa = 10.75SFLNTFYY378 pKa = 11.37NVLEE382 pKa = 4.39RR383 pKa = 11.84KK384 pKa = 9.35LPKK387 pKa = 10.55CNTICVWSPPSAGKK401 pKa = 10.0NFFFDD406 pKa = 3.69VYY408 pKa = 10.89LHH410 pKa = 6.09YY411 pKa = 11.21LMNYY415 pKa = 8.03GQLGIMNKK423 pKa = 8.69TNNFSLQEE431 pKa = 4.05ATSKK435 pKa = 10.72RR436 pKa = 11.84VLLWNEE442 pKa = 3.63PNYY445 pKa = 10.81EE446 pKa = 3.97DD447 pKa = 6.12AYY449 pKa = 10.07TDD451 pKa = 3.63TLKK454 pKa = 10.55MLTGGDD460 pKa = 3.89ALCVRR465 pKa = 11.84VKK467 pKa = 10.36QKK469 pKa = 10.85KK470 pKa = 9.01DD471 pKa = 2.96CHH473 pKa = 6.43VYY475 pKa = 8.51KK476 pKa = 10.51TPLIVLTNNMIGFMHH491 pKa = 7.01EE492 pKa = 3.82LAFVDD497 pKa = 3.87RR498 pKa = 11.84VKK500 pKa = 10.84VYY502 pKa = 9.69RR503 pKa = 11.84WKK505 pKa = 10.37QAPFLAEE512 pKa = 4.06YY513 pKa = 10.07NKK515 pKa = 10.57KK516 pKa = 10.12PNPLVAFEE524 pKa = 4.39ILVYY528 pKa = 10.11WEE530 pKa = 4.46IIPRR534 pKa = 11.84IIEE537 pKa = 3.79QQ538 pKa = 3.25
Molecular weight: 61.75 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2180 |
121 |
574 |
311.4 |
35.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.413 ± 0.567 | 1.972 ± 0.545 |
5.963 ± 0.573 | 6.239 ± 0.643 |
3.028 ± 0.336 | 5.78 ± 1.104 |
2.294 ± 0.301 | 6.376 ± 0.671 |
5.367 ± 0.426 | 8.349 ± 0.784 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.248 ± 0.345 | 5.459 ± 0.548 |
4.862 ± 0.732 | 4.679 ± 0.651 |
4.862 ± 0.719 | 7.523 ± 0.73 |
6.697 ± 0.583 | 6.697 ± 0.596 |
1.56 ± 0.448 | 4.633 ± 0.462 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
