
Gammapapillomavirus 8
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
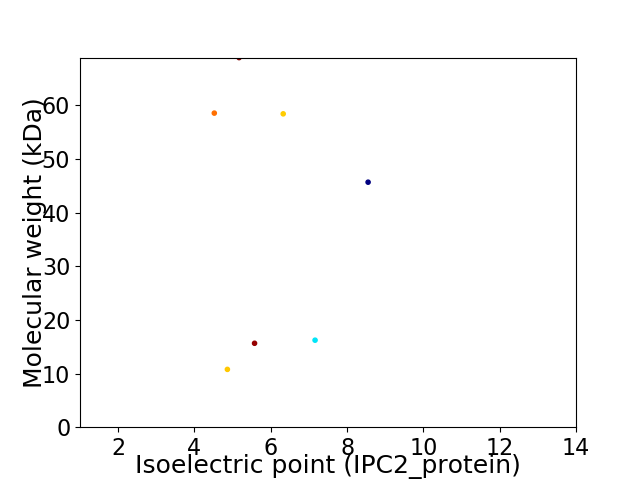
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2D2ALG0|A0A2D2ALG0_9PAPI Minor capsid protein L2 OS=Gammapapillomavirus 8 OX=1175850 GN=L2 PE=3 SV=1
MM1 pKa = 6.42YY2 pKa = 7.79TAKK5 pKa = 10.2RR6 pKa = 11.84VKK8 pKa = 9.97RR9 pKa = 11.84DD10 pKa = 3.33SPEE13 pKa = 3.67NIYY16 pKa = 10.12RR17 pKa = 11.84HH18 pKa = 5.92CKK20 pKa = 9.1IAGTCPPDD28 pKa = 3.44VEE30 pKa = 5.18NKK32 pKa = 9.9IEE34 pKa = 4.0QNTLADD40 pKa = 4.01RR41 pKa = 11.84LLKK44 pKa = 10.46IFGSIIYY51 pKa = 10.28LGGLGIGSGRR61 pKa = 11.84GTGAATGFRR70 pKa = 11.84PLPDD74 pKa = 3.99EE75 pKa = 4.84VPLPDD80 pKa = 3.81TNIITDD86 pKa = 3.51SSTLPEE92 pKa = 4.06TVRR95 pKa = 11.84PSLRR99 pKa = 11.84PRR101 pKa = 11.84PSRR104 pKa = 11.84PNTFGVPLDD113 pKa = 5.03PISSADD119 pKa = 3.6NIPRR123 pKa = 11.84IVRR126 pKa = 11.84PTEE129 pKa = 3.59PAIVPLSEE137 pKa = 5.1GGLPDD142 pKa = 3.46PTVISTGVEE151 pKa = 3.83SGAGVGDD158 pKa = 3.85YY159 pKa = 10.67EE160 pKa = 4.38VLTNIFVDD168 pKa = 3.97DD169 pKa = 3.89ALGAVGGHH177 pKa = 5.46PTVTSGVNEE186 pKa = 4.29NIALLEE192 pKa = 3.99ITPVEE197 pKa = 4.69HH198 pKa = 6.05IPSRR202 pKa = 11.84ITYY205 pKa = 6.96TTTDD209 pKa = 2.74VDD211 pKa = 3.23TGYY214 pKa = 8.87TFIRR218 pKa = 11.84SSLPADD224 pKa = 3.65PDD226 pKa = 3.55MNVFVDD232 pKa = 3.74PRR234 pKa = 11.84YY235 pKa = 10.16SGTVVGDD242 pKa = 3.86NIEE245 pKa = 4.4LEE247 pKa = 4.65AIHH250 pKa = 6.79NIDD253 pKa = 3.09QFEE256 pKa = 4.21IEE258 pKa = 4.65EE259 pKa = 4.35AGQPQTSTPSRR270 pKa = 11.84ISDD273 pKa = 3.51WAKK276 pKa = 10.41LRR278 pKa = 11.84AQEE281 pKa = 4.25FYY283 pKa = 11.49NRR285 pKa = 11.84FIQQMPTRR293 pKa = 11.84NIDD296 pKa = 3.63FLGQASRR303 pKa = 11.84AVQFEE308 pKa = 4.18FEE310 pKa = 4.24NPAFEE315 pKa = 5.03SDD317 pKa = 3.13VSLQFEE323 pKa = 4.09RR324 pKa = 11.84DD325 pKa = 3.77VQEE328 pKa = 4.14LAAAPDD334 pKa = 3.52EE335 pKa = 4.44AFTDD339 pKa = 3.89IVRR342 pKa = 11.84LGRR345 pKa = 11.84PVLSEE350 pKa = 4.12TPAGNVRR357 pKa = 11.84VSRR360 pKa = 11.84LGTRR364 pKa = 11.84GGMRR368 pKa = 11.84TRR370 pKa = 11.84SGTILKK376 pKa = 10.2QKK378 pKa = 9.57VHH380 pKa = 6.54FFYY383 pKa = 11.0DD384 pKa = 3.92LSPIAAVDD392 pKa = 3.68EE393 pKa = 4.55SAATNIEE400 pKa = 3.84LANFGEE406 pKa = 4.52SSDD409 pKa = 3.23ILTIVDD415 pKa = 3.96EE416 pKa = 4.62LSGGRR421 pKa = 11.84PINAFEE427 pKa = 4.79EE428 pKa = 4.34YY429 pKa = 10.76DD430 pKa = 3.56EE431 pKa = 4.43TALLNTDD438 pKa = 3.16EE439 pKa = 4.97ATFQNAHH446 pKa = 6.41LEE448 pKa = 4.38VYY450 pKa = 10.06TDD452 pKa = 4.64DD453 pKa = 5.28LEE455 pKa = 4.51EE456 pKa = 4.16PQIIPSLITDD466 pKa = 3.8VTPKK470 pKa = 10.77NFVNPYY476 pKa = 7.64ITDD479 pKa = 3.41ILVSYY484 pKa = 9.47PSIIDD489 pKa = 3.58NFPFTPIVPTTPLSPTVQIDD509 pKa = 3.58AFGSDD514 pKa = 4.44YY515 pKa = 11.04YY516 pKa = 11.08LHH518 pKa = 7.28PSLLRR523 pKa = 11.84RR524 pKa = 11.84KK525 pKa = 8.48KK526 pKa = 10.46RR527 pKa = 11.84KK528 pKa = 7.58WFEE531 pKa = 3.63MSS533 pKa = 2.85
MM1 pKa = 6.42YY2 pKa = 7.79TAKK5 pKa = 10.2RR6 pKa = 11.84VKK8 pKa = 9.97RR9 pKa = 11.84DD10 pKa = 3.33SPEE13 pKa = 3.67NIYY16 pKa = 10.12RR17 pKa = 11.84HH18 pKa = 5.92CKK20 pKa = 9.1IAGTCPPDD28 pKa = 3.44VEE30 pKa = 5.18NKK32 pKa = 9.9IEE34 pKa = 4.0QNTLADD40 pKa = 4.01RR41 pKa = 11.84LLKK44 pKa = 10.46IFGSIIYY51 pKa = 10.28LGGLGIGSGRR61 pKa = 11.84GTGAATGFRR70 pKa = 11.84PLPDD74 pKa = 3.99EE75 pKa = 4.84VPLPDD80 pKa = 3.81TNIITDD86 pKa = 3.51SSTLPEE92 pKa = 4.06TVRR95 pKa = 11.84PSLRR99 pKa = 11.84PRR101 pKa = 11.84PSRR104 pKa = 11.84PNTFGVPLDD113 pKa = 5.03PISSADD119 pKa = 3.6NIPRR123 pKa = 11.84IVRR126 pKa = 11.84PTEE129 pKa = 3.59PAIVPLSEE137 pKa = 5.1GGLPDD142 pKa = 3.46PTVISTGVEE151 pKa = 3.83SGAGVGDD158 pKa = 3.85YY159 pKa = 10.67EE160 pKa = 4.38VLTNIFVDD168 pKa = 3.97DD169 pKa = 3.89ALGAVGGHH177 pKa = 5.46PTVTSGVNEE186 pKa = 4.29NIALLEE192 pKa = 3.99ITPVEE197 pKa = 4.69HH198 pKa = 6.05IPSRR202 pKa = 11.84ITYY205 pKa = 6.96TTTDD209 pKa = 2.74VDD211 pKa = 3.23TGYY214 pKa = 8.87TFIRR218 pKa = 11.84SSLPADD224 pKa = 3.65PDD226 pKa = 3.55MNVFVDD232 pKa = 3.74PRR234 pKa = 11.84YY235 pKa = 10.16SGTVVGDD242 pKa = 3.86NIEE245 pKa = 4.4LEE247 pKa = 4.65AIHH250 pKa = 6.79NIDD253 pKa = 3.09QFEE256 pKa = 4.21IEE258 pKa = 4.65EE259 pKa = 4.35AGQPQTSTPSRR270 pKa = 11.84ISDD273 pKa = 3.51WAKK276 pKa = 10.41LRR278 pKa = 11.84AQEE281 pKa = 4.25FYY283 pKa = 11.49NRR285 pKa = 11.84FIQQMPTRR293 pKa = 11.84NIDD296 pKa = 3.63FLGQASRR303 pKa = 11.84AVQFEE308 pKa = 4.18FEE310 pKa = 4.24NPAFEE315 pKa = 5.03SDD317 pKa = 3.13VSLQFEE323 pKa = 4.09RR324 pKa = 11.84DD325 pKa = 3.77VQEE328 pKa = 4.14LAAAPDD334 pKa = 3.52EE335 pKa = 4.44AFTDD339 pKa = 3.89IVRR342 pKa = 11.84LGRR345 pKa = 11.84PVLSEE350 pKa = 4.12TPAGNVRR357 pKa = 11.84VSRR360 pKa = 11.84LGTRR364 pKa = 11.84GGMRR368 pKa = 11.84TRR370 pKa = 11.84SGTILKK376 pKa = 10.2QKK378 pKa = 9.57VHH380 pKa = 6.54FFYY383 pKa = 11.0DD384 pKa = 3.92LSPIAAVDD392 pKa = 3.68EE393 pKa = 4.55SAATNIEE400 pKa = 3.84LANFGEE406 pKa = 4.52SSDD409 pKa = 3.23ILTIVDD415 pKa = 3.96EE416 pKa = 4.62LSGGRR421 pKa = 11.84PINAFEE427 pKa = 4.79EE428 pKa = 4.34YY429 pKa = 10.76DD430 pKa = 3.56EE431 pKa = 4.43TALLNTDD438 pKa = 3.16EE439 pKa = 4.97ATFQNAHH446 pKa = 6.41LEE448 pKa = 4.38VYY450 pKa = 10.06TDD452 pKa = 4.64DD453 pKa = 5.28LEE455 pKa = 4.51EE456 pKa = 4.16PQIIPSLITDD466 pKa = 3.8VTPKK470 pKa = 10.77NFVNPYY476 pKa = 7.64ITDD479 pKa = 3.41ILVSYY484 pKa = 9.47PSIIDD489 pKa = 3.58NFPFTPIVPTTPLSPTVQIDD509 pKa = 3.58AFGSDD514 pKa = 4.44YY515 pKa = 11.04YY516 pKa = 11.08LHH518 pKa = 7.28PSLLRR523 pKa = 11.84RR524 pKa = 11.84KK525 pKa = 8.48KK526 pKa = 10.46RR527 pKa = 11.84KK528 pKa = 7.58WFEE531 pKa = 3.63MSS533 pKa = 2.85
Molecular weight: 58.54 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2D2ALE6|A0A2D2ALE6_9PAPI Protein E6 OS=Gammapapillomavirus 8 OX=1175850 GN=E6 PE=3 SV=1
MM1 pKa = 7.29ARR3 pKa = 11.84KK4 pKa = 9.9EE5 pKa = 4.03EE6 pKa = 4.28TQEE9 pKa = 3.98SLTEE13 pKa = 4.23RR14 pKa = 11.84FDD16 pKa = 4.04ALQEE20 pKa = 4.78AILTMIEE27 pKa = 4.09ANATDD32 pKa = 4.05LASHH36 pKa = 6.8LKK38 pKa = 9.06YY39 pKa = 9.38WDD41 pKa = 3.5YY42 pKa = 11.33VRR44 pKa = 11.84KK45 pKa = 10.18EE46 pKa = 4.0NVTLYY51 pKa = 10.2YY52 pKa = 10.58ARR54 pKa = 11.84KK55 pKa = 9.46CGYY58 pKa = 9.62NRR60 pKa = 11.84LGLQPTPTPAVSEE73 pKa = 4.46YY74 pKa = 10.35NAKK77 pKa = 9.88QAIKK81 pKa = 8.62MQMLIRR87 pKa = 11.84SLMTSQYY94 pKa = 11.37ADD96 pKa = 3.56EE97 pKa = 5.09HH98 pKa = 5.82WTLSDD103 pKa = 3.49ISAEE107 pKa = 4.35VINTQPRR114 pKa = 11.84DD115 pKa = 3.59CFKK118 pKa = 10.73KK119 pKa = 9.51QGYY122 pKa = 6.22TVEE125 pKa = 3.65VWYY128 pKa = 10.47DD129 pKa = 3.29NDD131 pKa = 3.41RR132 pKa = 11.84SKK134 pKa = 11.55AFPYY138 pKa = 9.83TNWTAIYY145 pKa = 9.96YY146 pKa = 9.87QDD148 pKa = 5.36EE149 pKa = 4.23NGNWHH154 pKa = 5.62KK155 pKa = 9.92TEE157 pKa = 4.05GQVDD161 pKa = 4.01ANGMYY166 pKa = 10.0YY167 pKa = 10.19KK168 pKa = 10.2EE169 pKa = 4.28VNGDD173 pKa = 3.34LVYY176 pKa = 10.35FHH178 pKa = 7.19LFGPDD183 pKa = 3.03AEE185 pKa = 5.15KK186 pKa = 11.11YY187 pKa = 10.45GSTGEE192 pKa = 4.07WTVHH196 pKa = 5.99FKK198 pKa = 10.86QSTIVSSSSSFAKK211 pKa = 10.43RR212 pKa = 11.84SPEE215 pKa = 3.49PLKK218 pKa = 10.69RR219 pKa = 11.84KK220 pKa = 9.49RR221 pKa = 11.84DD222 pKa = 3.57EE223 pKa = 4.57SSPSTSWDD231 pKa = 3.78TPSTTQDD238 pKa = 2.59IVEE241 pKa = 4.3RR242 pKa = 11.84RR243 pKa = 11.84SEE245 pKa = 4.04RR246 pKa = 11.84QTTVSSSSEE255 pKa = 3.64ANLRR259 pKa = 11.84PRR261 pKa = 11.84RR262 pKa = 11.84GGEE265 pKa = 3.72QGEE268 pKa = 4.61LPTTTKK274 pKa = 10.27RR275 pKa = 11.84RR276 pKa = 11.84RR277 pKa = 11.84LQSEE281 pKa = 4.44LSAVPTPDD289 pKa = 3.2QVGTRR294 pKa = 11.84HH295 pKa = 6.27RR296 pKa = 11.84SAPRR300 pKa = 11.84SNLTRR305 pKa = 11.84LQRR308 pKa = 11.84LQIEE312 pKa = 4.48SWDD315 pKa = 3.79PPILIVKK322 pKa = 7.33GHH324 pKa = 6.62PNSLKK329 pKa = 9.9CWRR332 pKa = 11.84NRR334 pKa = 11.84LRR336 pKa = 11.84STANSLLYY344 pKa = 10.11EE345 pKa = 4.18YY346 pKa = 11.07SSTVWKK352 pKa = 9.91WIGDD356 pKa = 3.81NNNLLGSRR364 pKa = 11.84MLLSFTSVTQRR375 pKa = 11.84NQFLKK380 pKa = 10.45VVKK383 pKa = 9.22PPKK386 pKa = 10.25GSVFALGSLDD396 pKa = 3.67SLL398 pKa = 4.35
MM1 pKa = 7.29ARR3 pKa = 11.84KK4 pKa = 9.9EE5 pKa = 4.03EE6 pKa = 4.28TQEE9 pKa = 3.98SLTEE13 pKa = 4.23RR14 pKa = 11.84FDD16 pKa = 4.04ALQEE20 pKa = 4.78AILTMIEE27 pKa = 4.09ANATDD32 pKa = 4.05LASHH36 pKa = 6.8LKK38 pKa = 9.06YY39 pKa = 9.38WDD41 pKa = 3.5YY42 pKa = 11.33VRR44 pKa = 11.84KK45 pKa = 10.18EE46 pKa = 4.0NVTLYY51 pKa = 10.2YY52 pKa = 10.58ARR54 pKa = 11.84KK55 pKa = 9.46CGYY58 pKa = 9.62NRR60 pKa = 11.84LGLQPTPTPAVSEE73 pKa = 4.46YY74 pKa = 10.35NAKK77 pKa = 9.88QAIKK81 pKa = 8.62MQMLIRR87 pKa = 11.84SLMTSQYY94 pKa = 11.37ADD96 pKa = 3.56EE97 pKa = 5.09HH98 pKa = 5.82WTLSDD103 pKa = 3.49ISAEE107 pKa = 4.35VINTQPRR114 pKa = 11.84DD115 pKa = 3.59CFKK118 pKa = 10.73KK119 pKa = 9.51QGYY122 pKa = 6.22TVEE125 pKa = 3.65VWYY128 pKa = 10.47DD129 pKa = 3.29NDD131 pKa = 3.41RR132 pKa = 11.84SKK134 pKa = 11.55AFPYY138 pKa = 9.83TNWTAIYY145 pKa = 9.96YY146 pKa = 9.87QDD148 pKa = 5.36EE149 pKa = 4.23NGNWHH154 pKa = 5.62KK155 pKa = 9.92TEE157 pKa = 4.05GQVDD161 pKa = 4.01ANGMYY166 pKa = 10.0YY167 pKa = 10.19KK168 pKa = 10.2EE169 pKa = 4.28VNGDD173 pKa = 3.34LVYY176 pKa = 10.35FHH178 pKa = 7.19LFGPDD183 pKa = 3.03AEE185 pKa = 5.15KK186 pKa = 11.11YY187 pKa = 10.45GSTGEE192 pKa = 4.07WTVHH196 pKa = 5.99FKK198 pKa = 10.86QSTIVSSSSSFAKK211 pKa = 10.43RR212 pKa = 11.84SPEE215 pKa = 3.49PLKK218 pKa = 10.69RR219 pKa = 11.84KK220 pKa = 9.49RR221 pKa = 11.84DD222 pKa = 3.57EE223 pKa = 4.57SSPSTSWDD231 pKa = 3.78TPSTTQDD238 pKa = 2.59IVEE241 pKa = 4.3RR242 pKa = 11.84RR243 pKa = 11.84SEE245 pKa = 4.04RR246 pKa = 11.84QTTVSSSSEE255 pKa = 3.64ANLRR259 pKa = 11.84PRR261 pKa = 11.84RR262 pKa = 11.84GGEE265 pKa = 3.72QGEE268 pKa = 4.61LPTTTKK274 pKa = 10.27RR275 pKa = 11.84RR276 pKa = 11.84RR277 pKa = 11.84LQSEE281 pKa = 4.44LSAVPTPDD289 pKa = 3.2QVGTRR294 pKa = 11.84HH295 pKa = 6.27RR296 pKa = 11.84SAPRR300 pKa = 11.84SNLTRR305 pKa = 11.84LQRR308 pKa = 11.84LQIEE312 pKa = 4.48SWDD315 pKa = 3.79PPILIVKK322 pKa = 7.33GHH324 pKa = 6.62PNSLKK329 pKa = 9.9CWRR332 pKa = 11.84NRR334 pKa = 11.84LRR336 pKa = 11.84STANSLLYY344 pKa = 10.11EE345 pKa = 4.18YY346 pKa = 11.07SSTVWKK352 pKa = 9.91WIGDD356 pKa = 3.81NNNLLGSRR364 pKa = 11.84MLLSFTSVTQRR375 pKa = 11.84NQFLKK380 pKa = 10.45VVKK383 pKa = 9.22PPKK386 pKa = 10.25GSVFALGSLDD396 pKa = 3.67SLL398 pKa = 4.35
Molecular weight: 45.67 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2421 |
97 |
605 |
345.9 |
39.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.204 ± 0.555 | 1.983 ± 0.576 |
6.444 ± 0.37 | 6.237 ± 0.565 |
4.75 ± 0.505 | 5.411 ± 0.466 |
1.983 ± 0.254 | 5.494 ± 0.803 |
5.328 ± 0.756 | 9.583 ± 0.721 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.528 ± 0.19 | 5.081 ± 0.337 |
5.989 ± 0.902 | 4.585 ± 0.575 |
5.37 ± 0.669 | 7.683 ± 0.596 |
6.939 ± 0.498 | 5.618 ± 0.544 |
1.363 ± 0.356 | 3.428 ± 0.377 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
