
Escherichia phage Qbeta
Taxonomy: Viruses; Riboviria; Orthornavirae; Lenarviricota; Leviviricetes; Norzivirales; Fiersviridae; Qubevirus; Qubevirus durum
Average proteome isoelectric point is 7.13
Get precalculated fractions of proteins
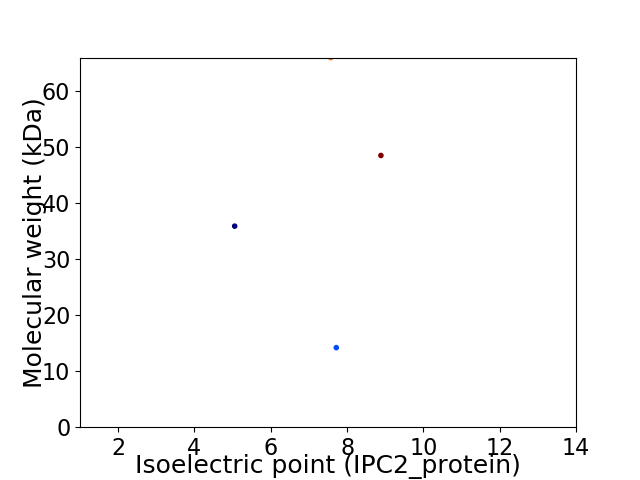
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
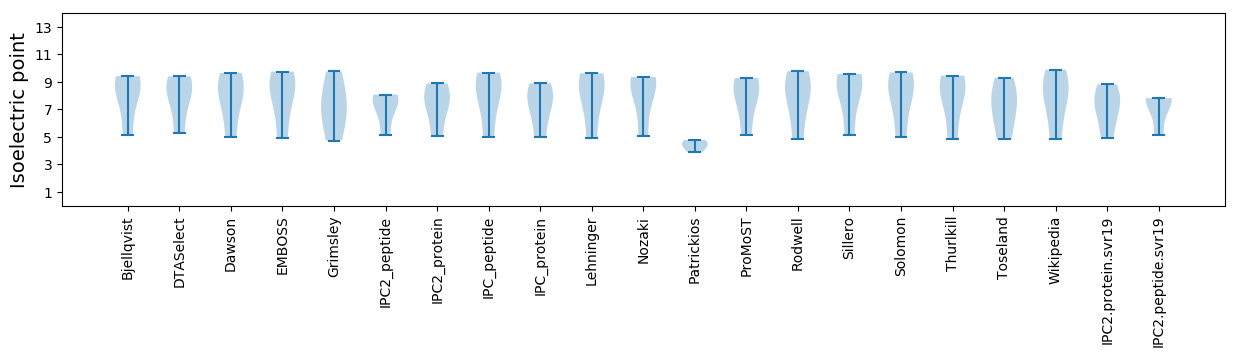
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|O64308|RDRP_BPMX1 RNA-directed RNA polymerase subunit beta OS=Escherichia phage Qbeta OX=2789016 PE=3 SV=1
MM1 pKa = 7.83AKK3 pKa = 9.99LQAITLSGIGKK14 pKa = 9.75NGDD17 pKa = 3.02VTLNLNPRR25 pKa = 11.84GVNPTNGVAALSEE38 pKa = 4.39AGAVPALEE46 pKa = 3.98KK47 pKa = 10.53RR48 pKa = 11.84VTISVSQPSRR58 pKa = 11.84NRR60 pKa = 11.84KK61 pKa = 8.01NYY63 pKa = 8.78KK64 pKa = 9.53VQVKK68 pKa = 9.25IQNPTSCTASGTCDD82 pKa = 3.32PSVTRR87 pKa = 11.84SAYY90 pKa = 10.58ADD92 pKa = 3.26VTFSFTQYY100 pKa = 9.75STDD103 pKa = 3.39EE104 pKa = 3.79EE105 pKa = 4.42RR106 pKa = 11.84ALVRR110 pKa = 11.84TEE112 pKa = 4.54LKK114 pKa = 10.83ALLADD119 pKa = 4.12PMLIDD124 pKa = 6.35AIDD127 pKa = 4.13NLNPAYY133 pKa = 7.22WTALLGDD140 pKa = 4.32GSGPSPVPGPNPDD153 pKa = 4.17PPLEE157 pKa = 4.47PPPGTGSYY165 pKa = 8.32TCPFRR170 pKa = 11.84IWDD173 pKa = 3.61LSSIYY178 pKa = 10.02EE179 pKa = 4.18AANSSHH185 pKa = 6.43SWDD188 pKa = 3.15IYY190 pKa = 10.68NAVEE194 pKa = 3.84LSPRR198 pKa = 11.84KK199 pKa = 9.52FDD201 pKa = 3.53VTLDD205 pKa = 3.87DD206 pKa = 5.62LLGNTDD212 pKa = 2.64WRR214 pKa = 11.84DD215 pKa = 2.46WDD217 pKa = 3.22GRR219 pKa = 11.84LRR221 pKa = 11.84YY222 pKa = 6.21TTFRR226 pKa = 11.84GSRR229 pKa = 11.84GNGYY233 pKa = 9.6IDD235 pKa = 5.51LDD237 pKa = 4.06ATSLMQDD244 pKa = 3.34EE245 pKa = 5.28YY246 pKa = 10.62LTSSKK251 pKa = 11.06YY252 pKa = 10.33LVRR255 pKa = 11.84EE256 pKa = 4.44GKK258 pKa = 10.38RR259 pKa = 11.84PGAFGSIEE267 pKa = 3.94RR268 pKa = 11.84FVYY271 pKa = 10.21LKK273 pKa = 10.67SINAYY278 pKa = 10.16CSLSDD283 pKa = 2.99ITAYY287 pKa = 10.59HH288 pKa = 6.45SDD290 pKa = 3.39GVVVGFWRR298 pKa = 11.84DD299 pKa = 3.36PSSGGAIPFDD309 pKa = 3.82FSEE312 pKa = 4.63FDD314 pKa = 3.73SNKK317 pKa = 9.97CPIQAVIVVPRR328 pKa = 11.84LL329 pKa = 3.22
MM1 pKa = 7.83AKK3 pKa = 9.99LQAITLSGIGKK14 pKa = 9.75NGDD17 pKa = 3.02VTLNLNPRR25 pKa = 11.84GVNPTNGVAALSEE38 pKa = 4.39AGAVPALEE46 pKa = 3.98KK47 pKa = 10.53RR48 pKa = 11.84VTISVSQPSRR58 pKa = 11.84NRR60 pKa = 11.84KK61 pKa = 8.01NYY63 pKa = 8.78KK64 pKa = 9.53VQVKK68 pKa = 9.25IQNPTSCTASGTCDD82 pKa = 3.32PSVTRR87 pKa = 11.84SAYY90 pKa = 10.58ADD92 pKa = 3.26VTFSFTQYY100 pKa = 9.75STDD103 pKa = 3.39EE104 pKa = 3.79EE105 pKa = 4.42RR106 pKa = 11.84ALVRR110 pKa = 11.84TEE112 pKa = 4.54LKK114 pKa = 10.83ALLADD119 pKa = 4.12PMLIDD124 pKa = 6.35AIDD127 pKa = 4.13NLNPAYY133 pKa = 7.22WTALLGDD140 pKa = 4.32GSGPSPVPGPNPDD153 pKa = 4.17PPLEE157 pKa = 4.47PPPGTGSYY165 pKa = 8.32TCPFRR170 pKa = 11.84IWDD173 pKa = 3.61LSSIYY178 pKa = 10.02EE179 pKa = 4.18AANSSHH185 pKa = 6.43SWDD188 pKa = 3.15IYY190 pKa = 10.68NAVEE194 pKa = 3.84LSPRR198 pKa = 11.84KK199 pKa = 9.52FDD201 pKa = 3.53VTLDD205 pKa = 3.87DD206 pKa = 5.62LLGNTDD212 pKa = 2.64WRR214 pKa = 11.84DD215 pKa = 2.46WDD217 pKa = 3.22GRR219 pKa = 11.84LRR221 pKa = 11.84YY222 pKa = 6.21TTFRR226 pKa = 11.84GSRR229 pKa = 11.84GNGYY233 pKa = 9.6IDD235 pKa = 5.51LDD237 pKa = 4.06ATSLMQDD244 pKa = 3.34EE245 pKa = 5.28YY246 pKa = 10.62LTSSKK251 pKa = 11.06YY252 pKa = 10.33LVRR255 pKa = 11.84EE256 pKa = 4.44GKK258 pKa = 10.38RR259 pKa = 11.84PGAFGSIEE267 pKa = 3.94RR268 pKa = 11.84FVYY271 pKa = 10.21LKK273 pKa = 10.67SINAYY278 pKa = 10.16CSLSDD283 pKa = 2.99ITAYY287 pKa = 10.59HH288 pKa = 6.45SDD290 pKa = 3.39GVVVGFWRR298 pKa = 11.84DD299 pKa = 3.36PSSGGAIPFDD309 pKa = 3.82FSEE312 pKa = 4.63FDD314 pKa = 3.73SNKK317 pKa = 9.97CPIQAVIVVPRR328 pKa = 11.84LL329 pKa = 3.22
Molecular weight: 35.88 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|O64307|A1_BPMX1 Minor capsid protein A1 OS=Escherichia phage Qbeta OX=2789016 PE=3 SV=1
MM1 pKa = 7.49PRR3 pKa = 11.84LPRR6 pKa = 11.84ALRR9 pKa = 11.84FGPNMEE15 pKa = 4.23VLSDD19 pKa = 4.32FQEE22 pKa = 3.67LWYY25 pKa = 9.69PEE27 pKa = 4.64SIIDD31 pKa = 4.36SDD33 pKa = 3.96VKK35 pKa = 10.95YY36 pKa = 10.31PLYY39 pKa = 10.23TFRR42 pKa = 11.84GSIGGSFFDD51 pKa = 5.17SYY53 pKa = 10.33GTNNIVRR60 pKa = 11.84EE61 pKa = 4.17IRR63 pKa = 11.84RR64 pKa = 11.84TPHH67 pKa = 6.52CATVPIASSGLRR79 pKa = 11.84PCTSVWYY86 pKa = 9.94DD87 pKa = 3.32PTSLLFRR94 pKa = 11.84IPEE97 pKa = 3.88MRR99 pKa = 11.84AEE101 pKa = 4.24WDD103 pKa = 3.2NGMGDD108 pKa = 4.07AGDD111 pKa = 3.49IVYY114 pKa = 10.64KK115 pKa = 10.77DD116 pKa = 4.03FLFSTPAPKK125 pKa = 10.6EE126 pKa = 3.76FDD128 pKa = 3.83FSNSLAPRR136 pKa = 11.84YY137 pKa = 10.3SNAFSAFNAKK147 pKa = 9.9YY148 pKa = 10.36GVIIGEE154 pKa = 4.02GHH156 pKa = 5.48EE157 pKa = 4.18TLKK160 pKa = 11.11YY161 pKa = 9.87FALLLRR167 pKa = 11.84RR168 pKa = 11.84LHH170 pKa = 5.78KK171 pKa = 10.13AVRR174 pKa = 11.84AVRR177 pKa = 11.84HH178 pKa = 5.48GDD180 pKa = 3.2LRR182 pKa = 11.84GLRR185 pKa = 11.84KK186 pKa = 9.57ILDD189 pKa = 3.91SYY191 pKa = 11.85NKK193 pKa = 9.9GRR195 pKa = 11.84WKK197 pKa = 10.18PATAGNLWLEE207 pKa = 4.14FRR209 pKa = 11.84YY210 pKa = 10.65GLTPLFHH217 pKa = 7.6DD218 pKa = 4.61IKK220 pKa = 11.2SVMDD224 pKa = 3.35DD225 pKa = 3.07WNRR228 pKa = 11.84INDD231 pKa = 4.36KK232 pKa = 10.01IQKK235 pKa = 9.24LRR237 pKa = 11.84RR238 pKa = 11.84FSVGHH243 pKa = 6.33GEE245 pKa = 4.18DD246 pKa = 3.63FKK248 pKa = 11.76LSIDD252 pKa = 3.54GLYY255 pKa = 10.28PGLTHH260 pKa = 6.94FRR262 pKa = 11.84LSGEE266 pKa = 3.67ITVQRR271 pKa = 11.84RR272 pKa = 11.84HH273 pKa = 5.28RR274 pKa = 11.84WGITYY279 pKa = 10.49ANRR282 pKa = 11.84EE283 pKa = 4.14GYY285 pKa = 8.46ATFDD289 pKa = 3.42NGSIRR294 pKa = 11.84PVSDD298 pKa = 2.51WKK300 pKa = 10.89EE301 pKa = 3.5LANAFINPGEE311 pKa = 4.46VAWEE315 pKa = 3.73LTPYY319 pKa = 10.82SFIVDD324 pKa = 2.97WFINVGDD331 pKa = 4.74IIEE334 pKa = 4.29QQKK337 pKa = 9.3QWYY340 pKa = 8.36QNIDD344 pKa = 3.11IVDD347 pKa = 4.76GYY349 pKa = 8.32QRR351 pKa = 11.84RR352 pKa = 11.84DD353 pKa = 2.52IRR355 pKa = 11.84MRR357 pKa = 11.84SVSLKK362 pKa = 9.88GVRR365 pKa = 11.84NGIPVRR371 pKa = 11.84VTGSVEE377 pKa = 4.25LVDD380 pKa = 3.9SFYY383 pKa = 11.43NRR385 pKa = 11.84SHH387 pKa = 5.49TTRR390 pKa = 11.84IPQATLAIDD399 pKa = 3.55TSFSSIKK406 pKa = 10.38HH407 pKa = 5.03VMDD410 pKa = 5.31SISLITQRR418 pKa = 11.84IKK420 pKa = 10.83RR421 pKa = 3.79
MM1 pKa = 7.49PRR3 pKa = 11.84LPRR6 pKa = 11.84ALRR9 pKa = 11.84FGPNMEE15 pKa = 4.23VLSDD19 pKa = 4.32FQEE22 pKa = 3.67LWYY25 pKa = 9.69PEE27 pKa = 4.64SIIDD31 pKa = 4.36SDD33 pKa = 3.96VKK35 pKa = 10.95YY36 pKa = 10.31PLYY39 pKa = 10.23TFRR42 pKa = 11.84GSIGGSFFDD51 pKa = 5.17SYY53 pKa = 10.33GTNNIVRR60 pKa = 11.84EE61 pKa = 4.17IRR63 pKa = 11.84RR64 pKa = 11.84TPHH67 pKa = 6.52CATVPIASSGLRR79 pKa = 11.84PCTSVWYY86 pKa = 9.94DD87 pKa = 3.32PTSLLFRR94 pKa = 11.84IPEE97 pKa = 3.88MRR99 pKa = 11.84AEE101 pKa = 4.24WDD103 pKa = 3.2NGMGDD108 pKa = 4.07AGDD111 pKa = 3.49IVYY114 pKa = 10.64KK115 pKa = 10.77DD116 pKa = 4.03FLFSTPAPKK125 pKa = 10.6EE126 pKa = 3.76FDD128 pKa = 3.83FSNSLAPRR136 pKa = 11.84YY137 pKa = 10.3SNAFSAFNAKK147 pKa = 9.9YY148 pKa = 10.36GVIIGEE154 pKa = 4.02GHH156 pKa = 5.48EE157 pKa = 4.18TLKK160 pKa = 11.11YY161 pKa = 9.87FALLLRR167 pKa = 11.84RR168 pKa = 11.84LHH170 pKa = 5.78KK171 pKa = 10.13AVRR174 pKa = 11.84AVRR177 pKa = 11.84HH178 pKa = 5.48GDD180 pKa = 3.2LRR182 pKa = 11.84GLRR185 pKa = 11.84KK186 pKa = 9.57ILDD189 pKa = 3.91SYY191 pKa = 11.85NKK193 pKa = 9.9GRR195 pKa = 11.84WKK197 pKa = 10.18PATAGNLWLEE207 pKa = 4.14FRR209 pKa = 11.84YY210 pKa = 10.65GLTPLFHH217 pKa = 7.6DD218 pKa = 4.61IKK220 pKa = 11.2SVMDD224 pKa = 3.35DD225 pKa = 3.07WNRR228 pKa = 11.84INDD231 pKa = 4.36KK232 pKa = 10.01IQKK235 pKa = 9.24LRR237 pKa = 11.84RR238 pKa = 11.84FSVGHH243 pKa = 6.33GEE245 pKa = 4.18DD246 pKa = 3.63FKK248 pKa = 11.76LSIDD252 pKa = 3.54GLYY255 pKa = 10.28PGLTHH260 pKa = 6.94FRR262 pKa = 11.84LSGEE266 pKa = 3.67ITVQRR271 pKa = 11.84RR272 pKa = 11.84HH273 pKa = 5.28RR274 pKa = 11.84WGITYY279 pKa = 10.49ANRR282 pKa = 11.84EE283 pKa = 4.14GYY285 pKa = 8.46ATFDD289 pKa = 3.42NGSIRR294 pKa = 11.84PVSDD298 pKa = 2.51WKK300 pKa = 10.89EE301 pKa = 3.5LANAFINPGEE311 pKa = 4.46VAWEE315 pKa = 3.73LTPYY319 pKa = 10.82SFIVDD324 pKa = 2.97WFINVGDD331 pKa = 4.74IIEE334 pKa = 4.29QQKK337 pKa = 9.3QWYY340 pKa = 8.36QNIDD344 pKa = 3.11IVDD347 pKa = 4.76GYY349 pKa = 8.32QRR351 pKa = 11.84RR352 pKa = 11.84DD353 pKa = 2.52IRR355 pKa = 11.84MRR357 pKa = 11.84SVSLKK362 pKa = 9.88GVRR365 pKa = 11.84NGIPVRR371 pKa = 11.84VTGSVEE377 pKa = 4.25LVDD380 pKa = 3.9SFYY383 pKa = 11.43NRR385 pKa = 11.84SHH387 pKa = 5.49TTRR390 pKa = 11.84IPQATLAIDD399 pKa = 3.55TSFSSIKK406 pKa = 10.38HH407 pKa = 5.03VMDD410 pKa = 5.31SISLITQRR418 pKa = 11.84IKK420 pKa = 10.83RR421 pKa = 3.79
Molecular weight: 48.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1469 |
133 |
586 |
367.3 |
41.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.739 ± 0.871 | 1.566 ± 0.43 |
6.399 ± 0.379 | 4.493 ± 0.39 |
4.493 ± 0.686 | 6.807 ± 0.356 |
1.361 ± 0.415 | 6.059 ± 0.654 |
4.289 ± 0.194 | 8.986 ± 0.523 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.498 ± 0.168 | 4.969 ± 0.43 |
5.582 ± 0.61 | 2.246 ± 0.257 |
7.42 ± 0.729 | 8.918 ± 0.417 |
5.718 ± 0.681 | 6.739 ± 0.401 |
1.77 ± 0.362 | 3.948 ± 0.184 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
