
Clostridiaceae bacterium OM08-6BH
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Clostridiaceae; unclassified Clostridiaceae
Average proteome isoelectric point is 6.02
Get precalculated fractions of proteins
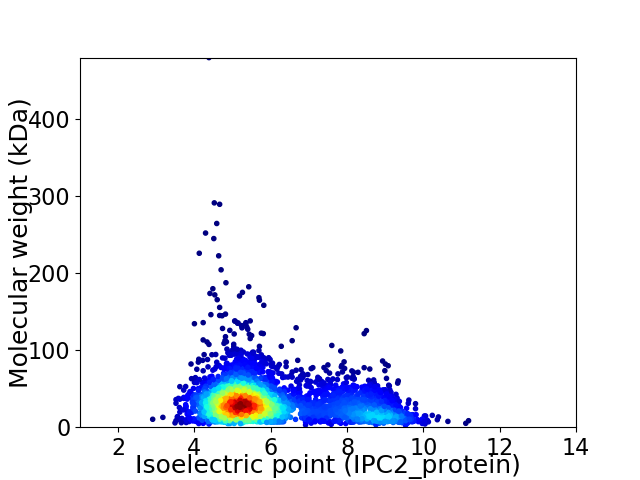
Virtual 2D-PAGE plot for 3400 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A417PSY0|A0A417PSY0_9CLOT Cell division protein FtsW OS=Clostridiaceae bacterium OM08-6BH OX=2292274 GN=DXC26_02850 PE=4 SV=1
MM1 pKa = 7.24KK2 pKa = 10.06RR3 pKa = 11.84KK4 pKa = 9.52LVALSLAVIMTAALTACGEE23 pKa = 4.2NSSSGTEE30 pKa = 4.01TQEE33 pKa = 4.1EE34 pKa = 4.82EE35 pKa = 4.23TTASEE40 pKa = 4.19EE41 pKa = 4.49TEE43 pKa = 4.31GTDD46 pKa = 3.31LQAAIVDD53 pKa = 3.36IDD55 pKa = 4.08EE56 pKa = 4.75APEE59 pKa = 3.96LTGSYY64 pKa = 10.08TIGLVIDD71 pKa = 4.01NNSDD75 pKa = 3.47QFNHH79 pKa = 6.5EE80 pKa = 4.25EE81 pKa = 4.32GEE83 pKa = 4.16AMKK86 pKa = 10.82ALGAAYY92 pKa = 10.17SSADD96 pKa = 3.33VTVTVMDD103 pKa = 3.98GQGDD107 pKa = 3.94VMKK110 pKa = 10.72QNQCVEE116 pKa = 3.98DD117 pKa = 4.76LINKK121 pKa = 9.1GVDD124 pKa = 4.03LILIIPIDD132 pKa = 3.85ADD134 pKa = 3.86GNVPAVNEE142 pKa = 3.99ALEE145 pKa = 4.2AGIPVVALDD154 pKa = 3.7ADD156 pKa = 4.33VNVEE160 pKa = 4.75DD161 pKa = 4.82DD162 pKa = 3.08MKK164 pKa = 10.9YY165 pKa = 10.18YY166 pKa = 10.92VGASDD171 pKa = 3.95YY172 pKa = 11.4NAGKK176 pKa = 9.91AQAEE180 pKa = 4.37YY181 pKa = 10.1LAEE184 pKa = 4.07VLPEE188 pKa = 3.94NANVLVILGQQGMSNAQNRR207 pKa = 11.84RR208 pKa = 11.84DD209 pKa = 3.71GAVEE213 pKa = 3.78TLTEE217 pKa = 4.16LRR219 pKa = 11.84PDD221 pKa = 3.35VTFLAEE227 pKa = 5.09QPGDD231 pKa = 3.38WDD233 pKa = 3.45EE234 pKa = 4.33AKK236 pKa = 10.79CMEE239 pKa = 5.0IVEE242 pKa = 4.86DD243 pKa = 3.37WCQAYY248 pKa = 10.04DD249 pKa = 3.44QFDD252 pKa = 4.61AIICGGDD259 pKa = 3.45SMALGAEE266 pKa = 4.31EE267 pKa = 5.12ALEE270 pKa = 3.99QADD273 pKa = 4.46RR274 pKa = 11.84LDD276 pKa = 3.42GVYY279 pKa = 8.65ITGVDD284 pKa = 4.41CIDD287 pKa = 3.53QVVALMKK294 pKa = 9.92EE295 pKa = 4.44GKK297 pKa = 9.51IAMSAQHH304 pKa = 5.92TADD307 pKa = 3.06MCAEE311 pKa = 4.01KK312 pKa = 10.73GFNYY316 pKa = 10.2ALHH319 pKa = 6.0VLNGEE324 pKa = 4.2NPDD327 pKa = 3.76DD328 pKa = 4.61YY329 pKa = 10.38IWEE332 pKa = 4.17YY333 pKa = 11.16VAVNKK338 pKa = 10.87DD339 pKa = 3.52NVNEE343 pKa = 4.63FYY345 pKa = 10.6PDD347 pKa = 2.98
MM1 pKa = 7.24KK2 pKa = 10.06RR3 pKa = 11.84KK4 pKa = 9.52LVALSLAVIMTAALTACGEE23 pKa = 4.2NSSSGTEE30 pKa = 4.01TQEE33 pKa = 4.1EE34 pKa = 4.82EE35 pKa = 4.23TTASEE40 pKa = 4.19EE41 pKa = 4.49TEE43 pKa = 4.31GTDD46 pKa = 3.31LQAAIVDD53 pKa = 3.36IDD55 pKa = 4.08EE56 pKa = 4.75APEE59 pKa = 3.96LTGSYY64 pKa = 10.08TIGLVIDD71 pKa = 4.01NNSDD75 pKa = 3.47QFNHH79 pKa = 6.5EE80 pKa = 4.25EE81 pKa = 4.32GEE83 pKa = 4.16AMKK86 pKa = 10.82ALGAAYY92 pKa = 10.17SSADD96 pKa = 3.33VTVTVMDD103 pKa = 3.98GQGDD107 pKa = 3.94VMKK110 pKa = 10.72QNQCVEE116 pKa = 3.98DD117 pKa = 4.76LINKK121 pKa = 9.1GVDD124 pKa = 4.03LILIIPIDD132 pKa = 3.85ADD134 pKa = 3.86GNVPAVNEE142 pKa = 3.99ALEE145 pKa = 4.2AGIPVVALDD154 pKa = 3.7ADD156 pKa = 4.33VNVEE160 pKa = 4.75DD161 pKa = 4.82DD162 pKa = 3.08MKK164 pKa = 10.9YY165 pKa = 10.18YY166 pKa = 10.92VGASDD171 pKa = 3.95YY172 pKa = 11.4NAGKK176 pKa = 9.91AQAEE180 pKa = 4.37YY181 pKa = 10.1LAEE184 pKa = 4.07VLPEE188 pKa = 3.94NANVLVILGQQGMSNAQNRR207 pKa = 11.84RR208 pKa = 11.84DD209 pKa = 3.71GAVEE213 pKa = 3.78TLTEE217 pKa = 4.16LRR219 pKa = 11.84PDD221 pKa = 3.35VTFLAEE227 pKa = 5.09QPGDD231 pKa = 3.38WDD233 pKa = 3.45EE234 pKa = 4.33AKK236 pKa = 10.79CMEE239 pKa = 5.0IVEE242 pKa = 4.86DD243 pKa = 3.37WCQAYY248 pKa = 10.04DD249 pKa = 3.44QFDD252 pKa = 4.61AIICGGDD259 pKa = 3.45SMALGAEE266 pKa = 4.31EE267 pKa = 5.12ALEE270 pKa = 3.99QADD273 pKa = 4.46RR274 pKa = 11.84LDD276 pKa = 3.42GVYY279 pKa = 8.65ITGVDD284 pKa = 4.41CIDD287 pKa = 3.53QVVALMKK294 pKa = 9.92EE295 pKa = 4.44GKK297 pKa = 9.51IAMSAQHH304 pKa = 5.92TADD307 pKa = 3.06MCAEE311 pKa = 4.01KK312 pKa = 10.73GFNYY316 pKa = 10.2ALHH319 pKa = 6.0VLNGEE324 pKa = 4.2NPDD327 pKa = 3.76DD328 pKa = 4.61YY329 pKa = 10.38IWEE332 pKa = 4.17YY333 pKa = 11.16VAVNKK338 pKa = 10.87DD339 pKa = 3.52NVNEE343 pKa = 4.63FYY345 pKa = 10.6PDD347 pKa = 2.98
Molecular weight: 37.36 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3R6SUC2|A0A3R6SUC2_9CLOT Uncharacterized protein OS=Clostridiaceae bacterium OM08-6BH OX=2292274 GN=DXC26_11380 PE=4 SV=1
MM1 pKa = 7.16RR2 pKa = 11.84TLRR5 pKa = 11.84QKK7 pKa = 10.37MSLARR12 pKa = 11.84DD13 pKa = 3.59QRR15 pKa = 11.84RR16 pKa = 11.84HH17 pKa = 5.31LSQNRR22 pKa = 11.84RR23 pKa = 11.84NQNQRR28 pKa = 11.84SQNRR32 pKa = 11.84RR33 pKa = 11.84NQNQRR38 pKa = 11.84NWNRR42 pKa = 11.84RR43 pKa = 11.84NQNQRR48 pKa = 11.84NRR50 pKa = 11.84NQRR53 pKa = 11.84NQNQRR58 pKa = 11.84NRR60 pKa = 11.84NWRR63 pKa = 11.84RR64 pKa = 2.92
MM1 pKa = 7.16RR2 pKa = 11.84TLRR5 pKa = 11.84QKK7 pKa = 10.37MSLARR12 pKa = 11.84DD13 pKa = 3.59QRR15 pKa = 11.84RR16 pKa = 11.84HH17 pKa = 5.31LSQNRR22 pKa = 11.84RR23 pKa = 11.84NQNQRR28 pKa = 11.84SQNRR32 pKa = 11.84RR33 pKa = 11.84NQNQRR38 pKa = 11.84NWNRR42 pKa = 11.84RR43 pKa = 11.84NQNQRR48 pKa = 11.84NRR50 pKa = 11.84NQRR53 pKa = 11.84NQNQRR58 pKa = 11.84NRR60 pKa = 11.84NWRR63 pKa = 11.84RR64 pKa = 2.92
Molecular weight: 8.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1057526 |
25 |
4441 |
311.0 |
34.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.68 ± 0.043 | 1.53 ± 0.017 |
5.643 ± 0.036 | 7.838 ± 0.048 |
3.942 ± 0.033 | 7.138 ± 0.044 |
1.833 ± 0.019 | 6.861 ± 0.036 |
6.608 ± 0.034 | 9.13 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.188 ± 0.027 | 4.06 ± 0.032 |
3.426 ± 0.024 | 3.479 ± 0.026 |
4.562 ± 0.046 | 5.634 ± 0.035 |
5.523 ± 0.052 | 6.866 ± 0.037 |
0.954 ± 0.016 | 4.104 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
