
Fusarium poae victorivirus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Totiviridae; Victorivirus; unclassified Victorivirus
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
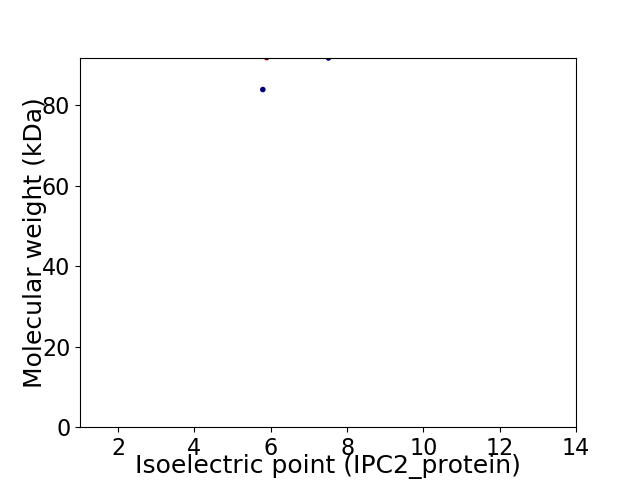
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
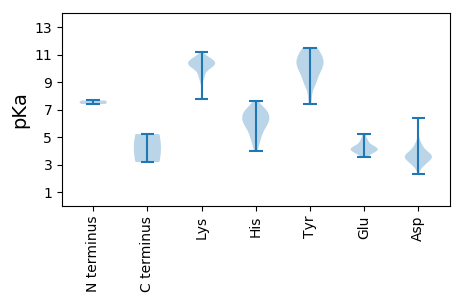
Protein with the lowest isoelectric point:
>tr|A0A1B4ZA57|A0A1B4ZA57_9VIRU RNA-directed RNA polymerase OS=Fusarium poae victorivirus 1 OX=1849535 PE=3 SV=1
MM1 pKa = 7.66PLSSPVFSSPAGRR14 pKa = 11.84VPLIRR19 pKa = 11.84PIEE22 pKa = 4.05LKK24 pKa = 10.76RR25 pKa = 11.84PINALFYY32 pKa = 10.97NSGAMAQPQSFRR44 pKa = 11.84TATTSALTGAVAGVSSGQIQDD65 pKa = 3.57DD66 pKa = 3.63NTYY69 pKa = 9.99RR70 pKa = 11.84RR71 pKa = 11.84YY72 pKa = 10.22RR73 pKa = 11.84SGLSIGVHH81 pKa = 4.39EE82 pKa = 5.24HH83 pKa = 4.0GHH85 pKa = 4.86YY86 pKa = 10.11AYY88 pKa = 9.7KK89 pKa = 10.29RR90 pKa = 11.84SSVFYY95 pKa = 9.97EE96 pKa = 3.63VGRR99 pKa = 11.84RR100 pKa = 11.84YY101 pKa = 10.48ARR103 pKa = 11.84LTQALAGRR111 pKa = 11.84PEE113 pKa = 4.18GGRR116 pKa = 11.84AGFDD120 pKa = 2.94ASVMVNPAEE129 pKa = 4.13AANFEE134 pKa = 3.9GWARR138 pKa = 11.84RR139 pKa = 11.84FSNFSPAWDD148 pKa = 3.65MMDD151 pKa = 3.23LAGVVEE157 pKa = 4.77RR158 pKa = 11.84LAKK161 pKa = 10.18AVAAQSVYY169 pKa = 11.18GGVSTTNMRR178 pKa = 11.84AGFPVSVVALGTLDD192 pKa = 3.79SPQTASSSSVFIPRR206 pKa = 11.84TVDD209 pKa = 3.13RR210 pKa = 11.84VGNDD214 pKa = 2.73NVFAVLAAAANGEE227 pKa = 4.38GATVTTDD234 pKa = 3.42VVRR237 pKa = 11.84LDD239 pKa = 3.42AATNQPVVPAVQGQALAQAAVEE261 pKa = 4.12GLRR264 pKa = 11.84ILGANFEE271 pKa = 4.35RR272 pKa = 11.84SGAGDD277 pKa = 3.57IFAYY281 pKa = 10.49ALTRR285 pKa = 11.84GIHH288 pKa = 5.32SVVSVVSHH296 pKa = 5.9TDD298 pKa = 2.71EE299 pKa = 5.02GGWLRR304 pKa = 11.84GVLRR308 pKa = 11.84RR309 pKa = 11.84TGFRR313 pKa = 11.84APYY316 pKa = 10.29GGINQDD322 pKa = 3.4LRR324 pKa = 11.84DD325 pKa = 4.04YY326 pKa = 10.64PQLPPLASLSTAATSAWVDD345 pKa = 4.32AIALKK350 pKa = 9.23TAAIVAHH357 pKa = 6.98CDD359 pKa = 2.99PCVRR363 pKa = 11.84ADD365 pKa = 3.18GGYY368 pKa = 8.88YY369 pKa = 9.98PSVFTASSGDD379 pKa = 3.22ISPPGTEE386 pKa = 3.71EE387 pKa = 4.72GEE389 pKa = 4.31VPEE392 pKa = 5.23AVATNIGRR400 pKa = 11.84QISSDD405 pKa = 3.31VGRR408 pKa = 11.84FAPLYY413 pKa = 9.82MRR415 pKa = 11.84GLCYY419 pKa = 10.46LFGMSSTSGIAEE431 pKa = 3.97AHH433 pKa = 6.27FSTTAADD440 pKa = 3.89HH441 pKa = 6.98LSGTTDD447 pKa = 2.29RR448 pKa = 11.84HH449 pKa = 5.06LRR451 pKa = 11.84HH452 pKa = 5.77RR453 pKa = 11.84TVAPYY458 pKa = 9.74FWIEE462 pKa = 3.95PTSLVPTNALGTTAEE477 pKa = 4.15AAGFGALTTAGVEE490 pKa = 3.88AAIPAFEE497 pKa = 4.11RR498 pKa = 11.84VRR500 pKa = 11.84EE501 pKa = 3.95IDD503 pKa = 3.22HH504 pKa = 5.95GHH506 pKa = 5.99NANFTTIGFKK516 pKa = 9.48MRR518 pKa = 11.84SARR521 pKa = 11.84TSGLVAAYY529 pKa = 8.95AANPAEE535 pKa = 4.14LSGLRR540 pKa = 11.84LYY542 pKa = 10.82QFDD545 pKa = 3.8EE546 pKa = 4.81SSVVLAGNNGPTNGDD561 pKa = 3.03VPAKK565 pKa = 10.46HH566 pKa = 6.1NAADD570 pKa = 4.06PLSSYY575 pKa = 10.7LWTRR579 pKa = 11.84GQSAIPAPAEE589 pKa = 3.65FMNIQGSYY597 pKa = 7.41AAKK600 pKa = 10.2YY601 pKa = 10.07KK602 pKa = 10.6VVDD605 pKa = 3.58WDD607 pKa = 3.99DD608 pKa = 4.25DD609 pKa = 4.56FNGTLGPLPEE619 pKa = 4.11AWEE622 pKa = 4.48LEE624 pKa = 4.18SHH626 pKa = 5.05PTKK629 pKa = 10.26WRR631 pKa = 11.84TSVPTALPAGASNYY645 pKa = 9.73ADD647 pKa = 3.04SGARR651 pKa = 11.84RR652 pKa = 11.84ARR654 pKa = 11.84SRR656 pKa = 11.84AGVALAQATLRR667 pKa = 11.84NRR669 pKa = 11.84GLGDD673 pKa = 3.54ANSPVISVSNVPPSWDD689 pKa = 3.56DD690 pKa = 3.28EE691 pKa = 4.61RR692 pKa = 11.84PATTRR697 pKa = 11.84LDD699 pKa = 3.55DD700 pKa = 3.52TRR702 pKa = 11.84TAEE705 pKa = 4.23HH706 pKa = 6.56NPAPGVVVTPGQGTDD721 pKa = 3.25TADD724 pKa = 3.72TPAHH728 pKa = 6.42LAAPALPIPHH738 pKa = 5.72QQPLRR743 pKa = 11.84GAPYY747 pKa = 9.35PPRR750 pKa = 11.84APGQLGGAQPPPPPVGPPAPPGPPGGPPNDD780 pKa = 5.42DD781 pKa = 4.84DD782 pKa = 4.56NQGPPPQAPPADD794 pKa = 3.8NHH796 pKa = 6.15PVPPAAA802 pKa = 5.21
MM1 pKa = 7.66PLSSPVFSSPAGRR14 pKa = 11.84VPLIRR19 pKa = 11.84PIEE22 pKa = 4.05LKK24 pKa = 10.76RR25 pKa = 11.84PINALFYY32 pKa = 10.97NSGAMAQPQSFRR44 pKa = 11.84TATTSALTGAVAGVSSGQIQDD65 pKa = 3.57DD66 pKa = 3.63NTYY69 pKa = 9.99RR70 pKa = 11.84RR71 pKa = 11.84YY72 pKa = 10.22RR73 pKa = 11.84SGLSIGVHH81 pKa = 4.39EE82 pKa = 5.24HH83 pKa = 4.0GHH85 pKa = 4.86YY86 pKa = 10.11AYY88 pKa = 9.7KK89 pKa = 10.29RR90 pKa = 11.84SSVFYY95 pKa = 9.97EE96 pKa = 3.63VGRR99 pKa = 11.84RR100 pKa = 11.84YY101 pKa = 10.48ARR103 pKa = 11.84LTQALAGRR111 pKa = 11.84PEE113 pKa = 4.18GGRR116 pKa = 11.84AGFDD120 pKa = 2.94ASVMVNPAEE129 pKa = 4.13AANFEE134 pKa = 3.9GWARR138 pKa = 11.84RR139 pKa = 11.84FSNFSPAWDD148 pKa = 3.65MMDD151 pKa = 3.23LAGVVEE157 pKa = 4.77RR158 pKa = 11.84LAKK161 pKa = 10.18AVAAQSVYY169 pKa = 11.18GGVSTTNMRR178 pKa = 11.84AGFPVSVVALGTLDD192 pKa = 3.79SPQTASSSSVFIPRR206 pKa = 11.84TVDD209 pKa = 3.13RR210 pKa = 11.84VGNDD214 pKa = 2.73NVFAVLAAAANGEE227 pKa = 4.38GATVTTDD234 pKa = 3.42VVRR237 pKa = 11.84LDD239 pKa = 3.42AATNQPVVPAVQGQALAQAAVEE261 pKa = 4.12GLRR264 pKa = 11.84ILGANFEE271 pKa = 4.35RR272 pKa = 11.84SGAGDD277 pKa = 3.57IFAYY281 pKa = 10.49ALTRR285 pKa = 11.84GIHH288 pKa = 5.32SVVSVVSHH296 pKa = 5.9TDD298 pKa = 2.71EE299 pKa = 5.02GGWLRR304 pKa = 11.84GVLRR308 pKa = 11.84RR309 pKa = 11.84TGFRR313 pKa = 11.84APYY316 pKa = 10.29GGINQDD322 pKa = 3.4LRR324 pKa = 11.84DD325 pKa = 4.04YY326 pKa = 10.64PQLPPLASLSTAATSAWVDD345 pKa = 4.32AIALKK350 pKa = 9.23TAAIVAHH357 pKa = 6.98CDD359 pKa = 2.99PCVRR363 pKa = 11.84ADD365 pKa = 3.18GGYY368 pKa = 8.88YY369 pKa = 9.98PSVFTASSGDD379 pKa = 3.22ISPPGTEE386 pKa = 3.71EE387 pKa = 4.72GEE389 pKa = 4.31VPEE392 pKa = 5.23AVATNIGRR400 pKa = 11.84QISSDD405 pKa = 3.31VGRR408 pKa = 11.84FAPLYY413 pKa = 9.82MRR415 pKa = 11.84GLCYY419 pKa = 10.46LFGMSSTSGIAEE431 pKa = 3.97AHH433 pKa = 6.27FSTTAADD440 pKa = 3.89HH441 pKa = 6.98LSGTTDD447 pKa = 2.29RR448 pKa = 11.84HH449 pKa = 5.06LRR451 pKa = 11.84HH452 pKa = 5.77RR453 pKa = 11.84TVAPYY458 pKa = 9.74FWIEE462 pKa = 3.95PTSLVPTNALGTTAEE477 pKa = 4.15AAGFGALTTAGVEE490 pKa = 3.88AAIPAFEE497 pKa = 4.11RR498 pKa = 11.84VRR500 pKa = 11.84EE501 pKa = 3.95IDD503 pKa = 3.22HH504 pKa = 5.95GHH506 pKa = 5.99NANFTTIGFKK516 pKa = 9.48MRR518 pKa = 11.84SARR521 pKa = 11.84TSGLVAAYY529 pKa = 8.95AANPAEE535 pKa = 4.14LSGLRR540 pKa = 11.84LYY542 pKa = 10.82QFDD545 pKa = 3.8EE546 pKa = 4.81SSVVLAGNNGPTNGDD561 pKa = 3.03VPAKK565 pKa = 10.46HH566 pKa = 6.1NAADD570 pKa = 4.06PLSSYY575 pKa = 10.7LWTRR579 pKa = 11.84GQSAIPAPAEE589 pKa = 3.65FMNIQGSYY597 pKa = 7.41AAKK600 pKa = 10.2YY601 pKa = 10.07KK602 pKa = 10.6VVDD605 pKa = 3.58WDD607 pKa = 3.99DD608 pKa = 4.25DD609 pKa = 4.56FNGTLGPLPEE619 pKa = 4.11AWEE622 pKa = 4.48LEE624 pKa = 4.18SHH626 pKa = 5.05PTKK629 pKa = 10.26WRR631 pKa = 11.84TSVPTALPAGASNYY645 pKa = 9.73ADD647 pKa = 3.04SGARR651 pKa = 11.84RR652 pKa = 11.84ARR654 pKa = 11.84SRR656 pKa = 11.84AGVALAQATLRR667 pKa = 11.84NRR669 pKa = 11.84GLGDD673 pKa = 3.54ANSPVISVSNVPPSWDD689 pKa = 3.56DD690 pKa = 3.28EE691 pKa = 4.61RR692 pKa = 11.84PATTRR697 pKa = 11.84LDD699 pKa = 3.55DD700 pKa = 3.52TRR702 pKa = 11.84TAEE705 pKa = 4.23HH706 pKa = 6.56NPAPGVVVTPGQGTDD721 pKa = 3.25TADD724 pKa = 3.72TPAHH728 pKa = 6.42LAAPALPIPHH738 pKa = 5.72QQPLRR743 pKa = 11.84GAPYY747 pKa = 9.35PPRR750 pKa = 11.84APGQLGGAQPPPPPVGPPAPPGPPGGPPNDD780 pKa = 5.42DD781 pKa = 4.84DD782 pKa = 4.56NQGPPPQAPPADD794 pKa = 3.8NHH796 pKa = 6.15PVPPAAA802 pKa = 5.21
Molecular weight: 83.9 kDa
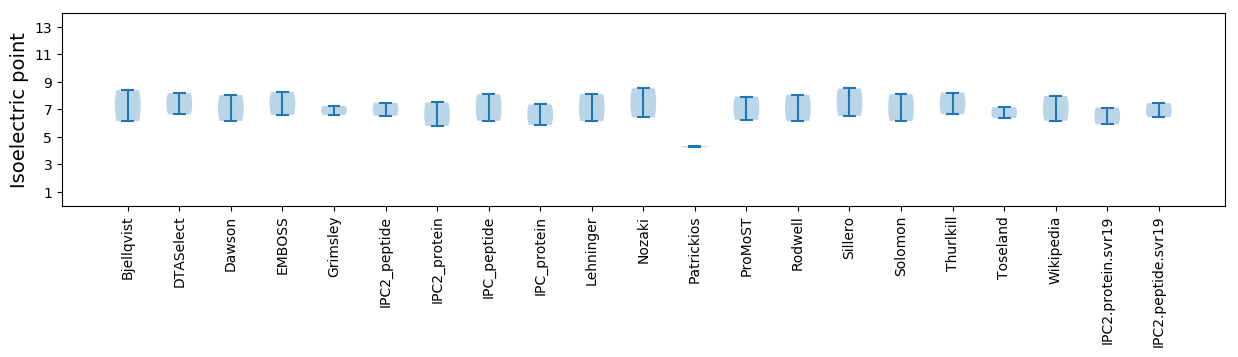
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B4ZA57|A0A1B4ZA57_9VIRU RNA-directed RNA polymerase OS=Fusarium poae victorivirus 1 OX=1849535 PE=3 SV=1
MM1 pKa = 7.41SPIDD5 pKa = 4.01RR6 pKa = 11.84GDD8 pKa = 3.55GEE10 pKa = 4.6AFLDD14 pKa = 4.31GVLDD18 pKa = 4.38SILKK22 pKa = 9.94RR23 pKa = 11.84FPGQLPDD30 pKa = 3.54VAGLPFDD37 pKa = 4.1EE38 pKa = 5.26QISAAYY44 pKa = 8.81SPSWGGVRR52 pKa = 11.84PSGLLRR58 pKa = 11.84AAFSYY63 pKa = 10.6RR64 pKa = 11.84HH65 pKa = 4.79TTVPVQSIYY74 pKa = 11.16DD75 pKa = 3.59HH76 pKa = 6.83SDD78 pKa = 2.59LRR80 pKa = 11.84RR81 pKa = 11.84LLNSVVEE88 pKa = 4.84FFPLSTDD95 pKa = 4.15FSSFKK100 pKa = 10.32ILKK103 pKa = 9.45HH104 pKa = 5.82PKK106 pKa = 9.85NSLKK110 pKa = 10.58LFKK113 pKa = 10.23PKK115 pKa = 10.35RR116 pKa = 11.84LPQALTKK123 pKa = 10.9ANLYY127 pKa = 10.08LDD129 pKa = 3.58EE130 pKa = 4.55VLRR133 pKa = 11.84DD134 pKa = 3.54LCKK137 pKa = 10.53NRR139 pKa = 11.84PTLGEE144 pKa = 3.99EE145 pKa = 3.97ASALLWSLRR154 pKa = 11.84DD155 pKa = 3.37RR156 pKa = 11.84GITHH160 pKa = 7.6DD161 pKa = 3.69AATAIVLYY169 pKa = 10.58GSALSQFYY177 pKa = 10.59TDD179 pKa = 3.59AFHH182 pKa = 6.83WAATAVLYY190 pKa = 9.26PKK192 pKa = 10.01LAKK195 pKa = 10.2AVSNFLKK202 pKa = 10.22ATGGNATSFGSLLVEE217 pKa = 4.43CEE219 pKa = 4.14VLQGRR224 pKa = 11.84GVGTIDD230 pKa = 4.35LLAEE234 pKa = 4.3AKK236 pKa = 10.09LRR238 pKa = 11.84CSPDD242 pKa = 3.29YY243 pKa = 11.46VRR245 pKa = 11.84DD246 pKa = 3.6HH247 pKa = 6.29YY248 pKa = 11.51AAAFDD253 pKa = 3.89EE254 pKa = 4.71EE255 pKa = 4.41KK256 pKa = 10.7LRR258 pKa = 11.84RR259 pKa = 11.84AVRR262 pKa = 11.84RR263 pKa = 11.84VYY265 pKa = 8.98EE266 pKa = 4.13TEE268 pKa = 3.77ISHH271 pKa = 6.85EE272 pKa = 4.4DD273 pKa = 3.16DD274 pKa = 3.43SQRR277 pKa = 11.84VEE279 pKa = 4.3FPTLEE284 pKa = 4.17EE285 pKa = 4.15HH286 pKa = 7.09WDD288 pKa = 3.86SRR290 pKa = 11.84WVWAVNGSQSSLLDD304 pKa = 3.3GGRR307 pKa = 11.84VKK309 pKa = 10.7KK310 pKa = 10.59LLEE313 pKa = 3.93PLGLHH318 pKa = 6.14KK319 pKa = 10.42LHH321 pKa = 6.54RR322 pKa = 11.84RR323 pKa = 11.84AWLEE327 pKa = 3.88CTPDD331 pKa = 3.69DD332 pKa = 5.34PRR334 pKa = 11.84VGWDD338 pKa = 2.85GTTYY342 pKa = 10.71VSPSPKK348 pKa = 10.18LEE350 pKa = 4.09NGKK353 pKa = 7.79TRR355 pKa = 11.84AIFACDD361 pKa = 3.12TRR363 pKa = 11.84HH364 pKa = 5.45YY365 pKa = 10.91LAFEE369 pKa = 4.27HH370 pKa = 7.0LLTPVEE376 pKa = 4.13KK377 pKa = 9.41RR378 pKa = 11.84WRR380 pKa = 11.84GSRR383 pKa = 11.84VVLNPGKK390 pKa = 10.53GGNIAMAQRR399 pKa = 11.84VRR401 pKa = 11.84HH402 pKa = 5.45SRR404 pKa = 11.84DD405 pKa = 2.85RR406 pKa = 11.84SGISLMLDD414 pKa = 2.95YY415 pKa = 11.45DD416 pKa = 4.37DD417 pKa = 6.41FNSQHH422 pKa = 5.4TTSAMKK428 pKa = 10.09IVIEE432 pKa = 4.15EE433 pKa = 4.25LCSVVGYY440 pKa = 8.94PADD443 pKa = 3.62LTATLVSSFDD453 pKa = 3.04KK454 pKa = 10.63MRR456 pKa = 11.84IHH458 pKa = 6.91VAGKK462 pKa = 9.61YY463 pKa = 9.83VGVAAGTLMSGHH475 pKa = 7.16RR476 pKa = 11.84ATTFINSVLNKK487 pKa = 10.25AYY489 pKa = 10.65LDD491 pKa = 3.82VVLGEE496 pKa = 4.26GWLDD500 pKa = 3.36TRR502 pKa = 11.84RR503 pKa = 11.84SVHH506 pKa = 6.49VGDD509 pKa = 6.07DD510 pKa = 3.7IYY512 pKa = 11.39CGVKK516 pKa = 9.99SYY518 pKa = 11.27RR519 pKa = 11.84DD520 pKa = 3.18AAYY523 pKa = 9.91VVHH526 pKa = 6.76QITSSPLRR534 pKa = 11.84MNPTKK539 pKa = 10.55QSVGHH544 pKa = 6.08VSTEE548 pKa = 3.8FLRR551 pKa = 11.84LATAGRR557 pKa = 11.84DD558 pKa = 3.29TYY560 pKa = 11.38GYY562 pKa = 8.51VARR565 pKa = 11.84SIASLISGNWVSDD578 pKa = 4.02RR579 pKa = 11.84IMNSYY584 pKa = 9.7EE585 pKa = 3.8ALTTMVASARR595 pKa = 11.84TLANRR600 pKa = 11.84ARR602 pKa = 11.84DD603 pKa = 3.59VTLPLLLEE611 pKa = 4.36SAVKK615 pKa = 10.23RR616 pKa = 11.84VLSKK620 pKa = 11.2DD621 pKa = 3.88CIDD624 pKa = 3.91DD625 pKa = 3.63RR626 pKa = 11.84VIRR629 pKa = 11.84RR630 pKa = 11.84LLLGEE635 pKa = 3.94VAINNGPTFSSSGSYY650 pKa = 9.44TRR652 pKa = 11.84VTVRR656 pKa = 11.84AEE658 pKa = 3.57YY659 pKa = 8.92TARR662 pKa = 11.84DD663 pKa = 3.49TVGRR667 pKa = 11.84PKK669 pKa = 10.53LPHH672 pKa = 6.15QSTSRR677 pKa = 11.84YY678 pKa = 7.74LTTNTTDD685 pKa = 4.56FEE687 pKa = 5.03CEE689 pKa = 4.0TLSQAGISPEE699 pKa = 3.69QTMIEE704 pKa = 4.09SSYY707 pKa = 11.39SKK709 pKa = 10.64SLQFGDD715 pKa = 4.72LYY717 pKa = 11.05FDD719 pKa = 3.76RR720 pKa = 11.84LVAGDD725 pKa = 4.07VEE727 pKa = 4.73STPARR732 pKa = 11.84GSVGVEE738 pKa = 3.66WLVHH742 pKa = 4.44TTPPKK747 pKa = 10.64GVLSQYY753 pKa = 10.58PLLVLVKK760 pKa = 10.35SRR762 pKa = 11.84LPEE765 pKa = 3.93YY766 pKa = 10.38VVRR769 pKa = 11.84DD770 pKa = 3.63AVRR773 pKa = 11.84RR774 pKa = 11.84AGGNPQCTDD783 pKa = 3.72LEE785 pKa = 4.36LEE787 pKa = 3.9AWGEE791 pKa = 4.0YY792 pKa = 9.17SHH794 pKa = 6.64GCIVNDD800 pKa = 3.16VLAYY804 pKa = 10.49SDD806 pKa = 3.53AAAFGKK812 pKa = 8.92RR813 pKa = 11.84TDD815 pKa = 3.81CSVLTSTHH823 pKa = 6.48RR824 pKa = 11.84YY825 pKa = 8.0YY826 pKa = 11.38VV827 pKa = 3.18
MM1 pKa = 7.41SPIDD5 pKa = 4.01RR6 pKa = 11.84GDD8 pKa = 3.55GEE10 pKa = 4.6AFLDD14 pKa = 4.31GVLDD18 pKa = 4.38SILKK22 pKa = 9.94RR23 pKa = 11.84FPGQLPDD30 pKa = 3.54VAGLPFDD37 pKa = 4.1EE38 pKa = 5.26QISAAYY44 pKa = 8.81SPSWGGVRR52 pKa = 11.84PSGLLRR58 pKa = 11.84AAFSYY63 pKa = 10.6RR64 pKa = 11.84HH65 pKa = 4.79TTVPVQSIYY74 pKa = 11.16DD75 pKa = 3.59HH76 pKa = 6.83SDD78 pKa = 2.59LRR80 pKa = 11.84RR81 pKa = 11.84LLNSVVEE88 pKa = 4.84FFPLSTDD95 pKa = 4.15FSSFKK100 pKa = 10.32ILKK103 pKa = 9.45HH104 pKa = 5.82PKK106 pKa = 9.85NSLKK110 pKa = 10.58LFKK113 pKa = 10.23PKK115 pKa = 10.35RR116 pKa = 11.84LPQALTKK123 pKa = 10.9ANLYY127 pKa = 10.08LDD129 pKa = 3.58EE130 pKa = 4.55VLRR133 pKa = 11.84DD134 pKa = 3.54LCKK137 pKa = 10.53NRR139 pKa = 11.84PTLGEE144 pKa = 3.99EE145 pKa = 3.97ASALLWSLRR154 pKa = 11.84DD155 pKa = 3.37RR156 pKa = 11.84GITHH160 pKa = 7.6DD161 pKa = 3.69AATAIVLYY169 pKa = 10.58GSALSQFYY177 pKa = 10.59TDD179 pKa = 3.59AFHH182 pKa = 6.83WAATAVLYY190 pKa = 9.26PKK192 pKa = 10.01LAKK195 pKa = 10.2AVSNFLKK202 pKa = 10.22ATGGNATSFGSLLVEE217 pKa = 4.43CEE219 pKa = 4.14VLQGRR224 pKa = 11.84GVGTIDD230 pKa = 4.35LLAEE234 pKa = 4.3AKK236 pKa = 10.09LRR238 pKa = 11.84CSPDD242 pKa = 3.29YY243 pKa = 11.46VRR245 pKa = 11.84DD246 pKa = 3.6HH247 pKa = 6.29YY248 pKa = 11.51AAAFDD253 pKa = 3.89EE254 pKa = 4.71EE255 pKa = 4.41KK256 pKa = 10.7LRR258 pKa = 11.84RR259 pKa = 11.84AVRR262 pKa = 11.84RR263 pKa = 11.84VYY265 pKa = 8.98EE266 pKa = 4.13TEE268 pKa = 3.77ISHH271 pKa = 6.85EE272 pKa = 4.4DD273 pKa = 3.16DD274 pKa = 3.43SQRR277 pKa = 11.84VEE279 pKa = 4.3FPTLEE284 pKa = 4.17EE285 pKa = 4.15HH286 pKa = 7.09WDD288 pKa = 3.86SRR290 pKa = 11.84WVWAVNGSQSSLLDD304 pKa = 3.3GGRR307 pKa = 11.84VKK309 pKa = 10.7KK310 pKa = 10.59LLEE313 pKa = 3.93PLGLHH318 pKa = 6.14KK319 pKa = 10.42LHH321 pKa = 6.54RR322 pKa = 11.84RR323 pKa = 11.84AWLEE327 pKa = 3.88CTPDD331 pKa = 3.69DD332 pKa = 5.34PRR334 pKa = 11.84VGWDD338 pKa = 2.85GTTYY342 pKa = 10.71VSPSPKK348 pKa = 10.18LEE350 pKa = 4.09NGKK353 pKa = 7.79TRR355 pKa = 11.84AIFACDD361 pKa = 3.12TRR363 pKa = 11.84HH364 pKa = 5.45YY365 pKa = 10.91LAFEE369 pKa = 4.27HH370 pKa = 7.0LLTPVEE376 pKa = 4.13KK377 pKa = 9.41RR378 pKa = 11.84WRR380 pKa = 11.84GSRR383 pKa = 11.84VVLNPGKK390 pKa = 10.53GGNIAMAQRR399 pKa = 11.84VRR401 pKa = 11.84HH402 pKa = 5.45SRR404 pKa = 11.84DD405 pKa = 2.85RR406 pKa = 11.84SGISLMLDD414 pKa = 2.95YY415 pKa = 11.45DD416 pKa = 4.37DD417 pKa = 6.41FNSQHH422 pKa = 5.4TTSAMKK428 pKa = 10.09IVIEE432 pKa = 4.15EE433 pKa = 4.25LCSVVGYY440 pKa = 8.94PADD443 pKa = 3.62LTATLVSSFDD453 pKa = 3.04KK454 pKa = 10.63MRR456 pKa = 11.84IHH458 pKa = 6.91VAGKK462 pKa = 9.61YY463 pKa = 9.83VGVAAGTLMSGHH475 pKa = 7.16RR476 pKa = 11.84ATTFINSVLNKK487 pKa = 10.25AYY489 pKa = 10.65LDD491 pKa = 3.82VVLGEE496 pKa = 4.26GWLDD500 pKa = 3.36TRR502 pKa = 11.84RR503 pKa = 11.84SVHH506 pKa = 6.49VGDD509 pKa = 6.07DD510 pKa = 3.7IYY512 pKa = 11.39CGVKK516 pKa = 9.99SYY518 pKa = 11.27RR519 pKa = 11.84DD520 pKa = 3.18AAYY523 pKa = 9.91VVHH526 pKa = 6.76QITSSPLRR534 pKa = 11.84MNPTKK539 pKa = 10.55QSVGHH544 pKa = 6.08VSTEE548 pKa = 3.8FLRR551 pKa = 11.84LATAGRR557 pKa = 11.84DD558 pKa = 3.29TYY560 pKa = 11.38GYY562 pKa = 8.51VARR565 pKa = 11.84SIASLISGNWVSDD578 pKa = 4.02RR579 pKa = 11.84IMNSYY584 pKa = 9.7EE585 pKa = 3.8ALTTMVASARR595 pKa = 11.84TLANRR600 pKa = 11.84ARR602 pKa = 11.84DD603 pKa = 3.59VTLPLLLEE611 pKa = 4.36SAVKK615 pKa = 10.23RR616 pKa = 11.84VLSKK620 pKa = 11.2DD621 pKa = 3.88CIDD624 pKa = 3.91DD625 pKa = 3.63RR626 pKa = 11.84VIRR629 pKa = 11.84RR630 pKa = 11.84LLLGEE635 pKa = 3.94VAINNGPTFSSSGSYY650 pKa = 9.44TRR652 pKa = 11.84VTVRR656 pKa = 11.84AEE658 pKa = 3.57YY659 pKa = 8.92TARR662 pKa = 11.84DD663 pKa = 3.49TVGRR667 pKa = 11.84PKK669 pKa = 10.53LPHH672 pKa = 6.15QSTSRR677 pKa = 11.84YY678 pKa = 7.74LTTNTTDD685 pKa = 4.56FEE687 pKa = 5.03CEE689 pKa = 4.0TLSQAGISPEE699 pKa = 3.69QTMIEE704 pKa = 4.09SSYY707 pKa = 11.39SKK709 pKa = 10.64SLQFGDD715 pKa = 4.72LYY717 pKa = 11.05FDD719 pKa = 3.76RR720 pKa = 11.84LVAGDD725 pKa = 4.07VEE727 pKa = 4.73STPARR732 pKa = 11.84GSVGVEE738 pKa = 3.66WLVHH742 pKa = 4.44TTPPKK747 pKa = 10.64GVLSQYY753 pKa = 10.58PLLVLVKK760 pKa = 10.35SRR762 pKa = 11.84LPEE765 pKa = 3.93YY766 pKa = 10.38VVRR769 pKa = 11.84DD770 pKa = 3.63AVRR773 pKa = 11.84RR774 pKa = 11.84AGGNPQCTDD783 pKa = 3.72LEE785 pKa = 4.36LEE787 pKa = 3.9AWGEE791 pKa = 4.0YY792 pKa = 9.17SHH794 pKa = 6.64GCIVNDD800 pKa = 3.16VLAYY804 pKa = 10.49SDD806 pKa = 3.53AAAFGKK812 pKa = 8.92RR813 pKa = 11.84TDD815 pKa = 3.81CSVLTSTHH823 pKa = 6.48RR824 pKa = 11.84YY825 pKa = 8.0YY826 pKa = 11.38VV827 pKa = 3.18
Molecular weight: 91.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1629 |
802 |
827 |
814.5 |
87.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.357 ± 2.279 | 0.921 ± 0.385 |
5.77 ± 0.464 | 4.236 ± 0.437 |
3.192 ± 0.035 | 8.226 ± 0.97 |
2.517 ± 0.192 | 3.131 ± 0.097 |
2.64 ± 1.07 | 8.717 ± 1.575 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.228 ± 0.013 | 3.315 ± 0.476 |
6.937 ± 1.878 | 2.64 ± 0.337 |
7.182 ± 0.405 | 8.349 ± 0.524 |
6.753 ± 0.014 | 8.042 ± 0.307 |
1.412 ± 0.116 | 3.438 ± 0.402 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
