
Pleurocapsa sp. CCALA 161
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Pleurocapsales; Hyellaceae; Pleurocapsa; unclassified Pleurocapsa
Average proteome isoelectric point is 6.29
Get precalculated fractions of proteins
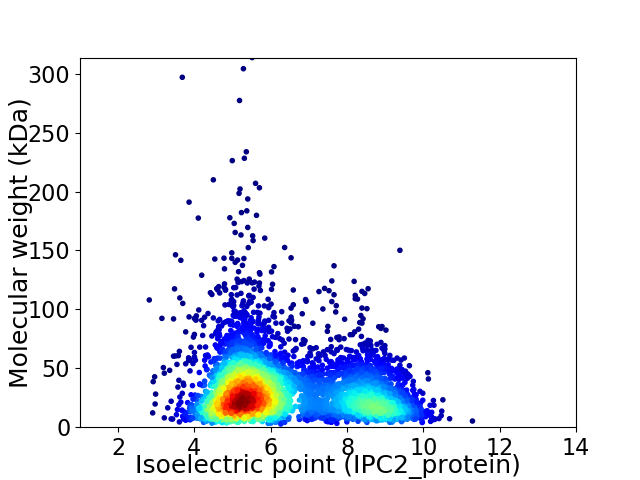
Virtual 2D-PAGE plot for 4800 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
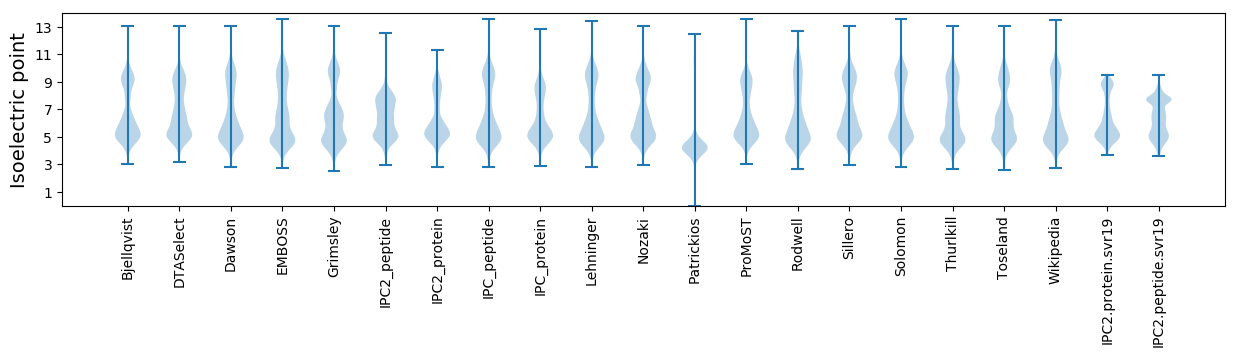
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2T1CQ04|A0A2T1CQ04_9CYAN Antitoxin OS=Pleurocapsa sp. CCALA 161 OX=2107688 GN=C7B62_09835 PE=3 SV=1
MM1 pKa = 7.73AFNTTSTIAEE11 pKa = 4.29DD12 pKa = 3.26WNGGYY17 pKa = 10.39KK18 pKa = 10.31LEE20 pKa = 4.31LAISNDD26 pKa = 3.07LGAADD31 pKa = 3.31WVLNFNLPYY40 pKa = 9.71TISAAYY46 pKa = 9.32GVNLTQNSDD55 pKa = 2.69GSYY58 pKa = 9.62TIKK61 pKa = 10.98GQDD64 pKa = 3.02GWTSLTPGQTIKK76 pKa = 10.72PIFIIEE82 pKa = 4.25DD83 pKa = 3.4QGQSAQLLNFVDD95 pKa = 4.19QQQSNPIATVDD106 pKa = 3.52DD107 pKa = 4.61FEE109 pKa = 7.34NIDD112 pKa = 3.64NSYY115 pKa = 10.72LPSGNNTIDD124 pKa = 3.04VDD126 pKa = 3.97RR127 pKa = 11.84DD128 pKa = 3.68FGGDD132 pKa = 3.29LATAIAYY139 pKa = 8.63ANNGDD144 pKa = 4.03VVQLGNNRR152 pKa = 11.84YY153 pKa = 6.29YY154 pKa = 10.88TSGITIDD161 pKa = 3.99KK162 pKa = 10.79NIILDD167 pKa = 3.86GQEE170 pKa = 4.27DD171 pKa = 4.45SVIDD175 pKa = 4.05GNGTYY180 pKa = 10.95NSIITLYY187 pKa = 10.38SGASGATIQDD197 pKa = 3.38IEE199 pKa = 4.4ITNGNNGIYY208 pKa = 10.15GYY210 pKa = 10.61KK211 pKa = 10.04AANLTLQNLEE221 pKa = 3.72VHH223 pKa = 6.66NIGKK227 pKa = 9.72NGMIADD233 pKa = 4.67GNQTGIAMDD242 pKa = 3.86HH243 pKa = 6.88ADD245 pKa = 4.05GLKK248 pKa = 10.53LSNTSVYY255 pKa = 10.57DD256 pKa = 3.46VEE258 pKa = 4.56RR259 pKa = 11.84KK260 pKa = 10.27GIGVGDD266 pKa = 3.74TNGAQITNVNVQSVNLAANHH286 pKa = 5.83AQSHH290 pKa = 6.36DD291 pKa = 3.32AAGIKK296 pKa = 9.77FFNTNNVILKK306 pKa = 10.19DD307 pKa = 3.7SYY309 pKa = 11.16FEE311 pKa = 4.87KK312 pKa = 10.7INAFYY317 pKa = 10.43IWNDD321 pKa = 3.41TTNKK325 pKa = 7.46TTIDD329 pKa = 3.58NNNVVNVGSDD339 pKa = 3.39FLAPSFNTNVNVAGIYY355 pKa = 9.44NEE357 pKa = 4.15KK358 pKa = 10.33SSNSIVRR365 pKa = 11.84YY366 pKa = 8.59NQGTAVGDD374 pKa = 4.33FSAFNATEE382 pKa = 4.64FSTQTMTFEE391 pKa = 5.28DD392 pKa = 3.87NNSFSKK398 pKa = 10.44MEE400 pKa = 5.14FGTQDD405 pKa = 2.9YY406 pKa = 8.5WVNQEE411 pKa = 4.08AEE413 pKa = 4.16MAIATTEE420 pKa = 4.11NPDD423 pKa = 3.3EE424 pKa = 4.84ANFSLFAAEE433 pKa = 4.8YY434 pKa = 8.43YY435 pKa = 10.18AQAVIQQ441 pKa = 3.78
MM1 pKa = 7.73AFNTTSTIAEE11 pKa = 4.29DD12 pKa = 3.26WNGGYY17 pKa = 10.39KK18 pKa = 10.31LEE20 pKa = 4.31LAISNDD26 pKa = 3.07LGAADD31 pKa = 3.31WVLNFNLPYY40 pKa = 9.71TISAAYY46 pKa = 9.32GVNLTQNSDD55 pKa = 2.69GSYY58 pKa = 9.62TIKK61 pKa = 10.98GQDD64 pKa = 3.02GWTSLTPGQTIKK76 pKa = 10.72PIFIIEE82 pKa = 4.25DD83 pKa = 3.4QGQSAQLLNFVDD95 pKa = 4.19QQQSNPIATVDD106 pKa = 3.52DD107 pKa = 4.61FEE109 pKa = 7.34NIDD112 pKa = 3.64NSYY115 pKa = 10.72LPSGNNTIDD124 pKa = 3.04VDD126 pKa = 3.97RR127 pKa = 11.84DD128 pKa = 3.68FGGDD132 pKa = 3.29LATAIAYY139 pKa = 8.63ANNGDD144 pKa = 4.03VVQLGNNRR152 pKa = 11.84YY153 pKa = 6.29YY154 pKa = 10.88TSGITIDD161 pKa = 3.99KK162 pKa = 10.79NIILDD167 pKa = 3.86GQEE170 pKa = 4.27DD171 pKa = 4.45SVIDD175 pKa = 4.05GNGTYY180 pKa = 10.95NSIITLYY187 pKa = 10.38SGASGATIQDD197 pKa = 3.38IEE199 pKa = 4.4ITNGNNGIYY208 pKa = 10.15GYY210 pKa = 10.61KK211 pKa = 10.04AANLTLQNLEE221 pKa = 3.72VHH223 pKa = 6.66NIGKK227 pKa = 9.72NGMIADD233 pKa = 4.67GNQTGIAMDD242 pKa = 3.86HH243 pKa = 6.88ADD245 pKa = 4.05GLKK248 pKa = 10.53LSNTSVYY255 pKa = 10.57DD256 pKa = 3.46VEE258 pKa = 4.56RR259 pKa = 11.84KK260 pKa = 10.27GIGVGDD266 pKa = 3.74TNGAQITNVNVQSVNLAANHH286 pKa = 5.83AQSHH290 pKa = 6.36DD291 pKa = 3.32AAGIKK296 pKa = 9.77FFNTNNVILKK306 pKa = 10.19DD307 pKa = 3.7SYY309 pKa = 11.16FEE311 pKa = 4.87KK312 pKa = 10.7INAFYY317 pKa = 10.43IWNDD321 pKa = 3.41TTNKK325 pKa = 7.46TTIDD329 pKa = 3.58NNNVVNVGSDD339 pKa = 3.39FLAPSFNTNVNVAGIYY355 pKa = 9.44NEE357 pKa = 4.15KK358 pKa = 10.33SSNSIVRR365 pKa = 11.84YY366 pKa = 8.59NQGTAVGDD374 pKa = 4.33FSAFNATEE382 pKa = 4.64FSTQTMTFEE391 pKa = 5.28DD392 pKa = 3.87NNSFSKK398 pKa = 10.44MEE400 pKa = 5.14FGTQDD405 pKa = 2.9YY406 pKa = 8.5WVNQEE411 pKa = 4.08AEE413 pKa = 4.16MAIATTEE420 pKa = 4.11NPDD423 pKa = 3.3EE424 pKa = 4.84ANFSLFAAEE433 pKa = 4.8YY434 pKa = 8.43YY435 pKa = 10.18AQAVIQQ441 pKa = 3.78
Molecular weight: 47.8 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2T1CFH2|A0A2T1CFH2_9CYAN NAD(P)-dependent oxidoreductase OS=Pleurocapsa sp. CCALA 161 OX=2107688 GN=C7B62_21305 PE=4 SV=1
MM1 pKa = 6.61TQRR4 pKa = 11.84TLGGTSRR11 pKa = 11.84KK12 pKa = 7.83QKK14 pKa = 9.77RR15 pKa = 11.84KK16 pKa = 9.16SGFRR20 pKa = 11.84ARR22 pKa = 11.84KK23 pKa = 7.01RR24 pKa = 11.84TSNGRR29 pKa = 11.84RR30 pKa = 11.84VISARR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.8KK38 pKa = 10.1GRR40 pKa = 11.84HH41 pKa = 5.0RR42 pKa = 11.84LSVV45 pKa = 3.12
MM1 pKa = 6.61TQRR4 pKa = 11.84TLGGTSRR11 pKa = 11.84KK12 pKa = 7.83QKK14 pKa = 9.77RR15 pKa = 11.84KK16 pKa = 9.16SGFRR20 pKa = 11.84ARR22 pKa = 11.84KK23 pKa = 7.01RR24 pKa = 11.84TSNGRR29 pKa = 11.84RR30 pKa = 11.84VISARR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.8KK38 pKa = 10.1GRR40 pKa = 11.84HH41 pKa = 5.0RR42 pKa = 11.84LSVV45 pKa = 3.12
Molecular weight: 5.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1462054 |
23 |
2812 |
304.6 |
33.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.042 ± 0.035 | 0.999 ± 0.012 |
5.118 ± 0.034 | 6.204 ± 0.032 |
3.897 ± 0.024 | 6.405 ± 0.036 |
1.781 ± 0.019 | 7.355 ± 0.032 |
5.246 ± 0.026 | 11.022 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.847 ± 0.015 | 4.67 ± 0.032 |
4.264 ± 0.026 | 5.523 ± 0.044 |
4.64 ± 0.024 | 6.617 ± 0.026 |
5.542 ± 0.028 | 6.279 ± 0.029 |
1.37 ± 0.016 | 3.178 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
