
Blackfly microvirus SF02
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae; Microvirus
Average proteome isoelectric point is 7.09
Get precalculated fractions of proteins
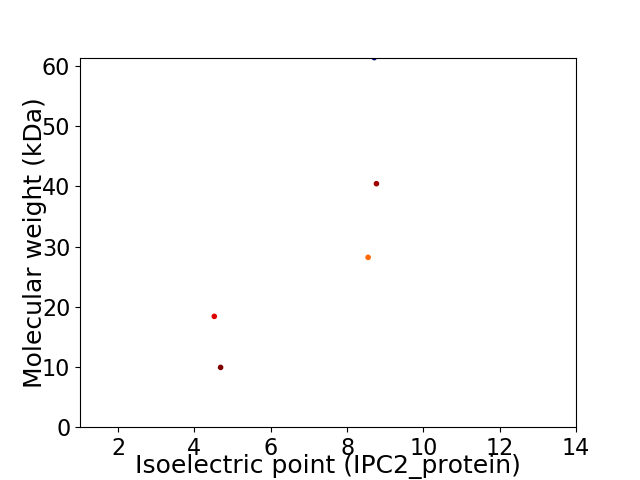
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A4P8PK03|A0A4P8PK03_9VIRU Major capsid protein OS=Blackfly microvirus SF02 OX=2576452 PE=3 SV=1
MM1 pKa = 7.37EE2 pKa = 4.66VNMSKK7 pKa = 10.49VSRR10 pKa = 11.84FQGAQAEE17 pKa = 4.24NDD19 pKa = 3.59FFKK22 pKa = 11.42SNGSDD27 pKa = 2.93TAVFNRR33 pKa = 11.84EE34 pKa = 3.53PSLTRR39 pKa = 11.84QEE41 pKa = 4.17FAEE44 pKa = 4.38EE45 pKa = 3.91CDD47 pKa = 3.24INQLMKK53 pKa = 10.53RR54 pKa = 11.84YY55 pKa = 8.85DD56 pKa = 3.29GHH58 pKa = 8.35VIGGPGNLPVRR69 pKa = 11.84EE70 pKa = 3.86PMYY73 pKa = 10.86FDD75 pKa = 5.44FADD78 pKa = 4.47LPQDD82 pKa = 3.28LMGYY86 pKa = 7.97LTFMNEE92 pKa = 3.33AQAQFMSLPAAVRR105 pKa = 11.84KK106 pKa = 9.92EE107 pKa = 3.77FDD109 pKa = 3.3NDD111 pKa = 3.26PVEE114 pKa = 4.72FVAYY118 pKa = 10.16ASDD121 pKa = 3.97PSSLEE126 pKa = 3.74QMRR129 pKa = 11.84SWGLAPPAKK138 pKa = 9.97PPEE141 pKa = 4.54TPVPPPPGEE150 pKa = 4.55GPASASKK157 pKa = 10.74GSSEE161 pKa = 4.08PPTHH165 pKa = 7.0GSTT168 pKa = 3.97
MM1 pKa = 7.37EE2 pKa = 4.66VNMSKK7 pKa = 10.49VSRR10 pKa = 11.84FQGAQAEE17 pKa = 4.24NDD19 pKa = 3.59FFKK22 pKa = 11.42SNGSDD27 pKa = 2.93TAVFNRR33 pKa = 11.84EE34 pKa = 3.53PSLTRR39 pKa = 11.84QEE41 pKa = 4.17FAEE44 pKa = 4.38EE45 pKa = 3.91CDD47 pKa = 3.24INQLMKK53 pKa = 10.53RR54 pKa = 11.84YY55 pKa = 8.85DD56 pKa = 3.29GHH58 pKa = 8.35VIGGPGNLPVRR69 pKa = 11.84EE70 pKa = 3.86PMYY73 pKa = 10.86FDD75 pKa = 5.44FADD78 pKa = 4.47LPQDD82 pKa = 3.28LMGYY86 pKa = 7.97LTFMNEE92 pKa = 3.33AQAQFMSLPAAVRR105 pKa = 11.84KK106 pKa = 9.92EE107 pKa = 3.77FDD109 pKa = 3.3NDD111 pKa = 3.26PVEE114 pKa = 4.72FVAYY118 pKa = 10.16ASDD121 pKa = 3.97PSSLEE126 pKa = 3.74QMRR129 pKa = 11.84SWGLAPPAKK138 pKa = 9.97PPEE141 pKa = 4.54TPVPPPPGEE150 pKa = 4.55GPASASKK157 pKa = 10.74GSSEE161 pKa = 4.08PPTHH165 pKa = 7.0GSTT168 pKa = 3.97
Molecular weight: 18.4 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4V1F5E5|A0A4V1F5E5_9VIRU Nonstructural protein OS=Blackfly microvirus SF02 OX=2576452 PE=4 SV=1
MM1 pKa = 7.9RR2 pKa = 11.84CHH4 pKa = 5.3QAVVSVHH11 pKa = 6.02LVRR14 pKa = 11.84ALVILIRR21 pKa = 11.84RR22 pKa = 11.84IFLLVSQCVAVFGCKK37 pKa = 8.95MGCVKK42 pKa = 10.11PLKK45 pKa = 10.36AFWRR49 pKa = 11.84SRR51 pKa = 11.84SRR53 pKa = 11.84DD54 pKa = 3.5AITFDD59 pKa = 3.35INKK62 pKa = 8.99SATRR66 pKa = 11.84ISFSLPCGRR75 pKa = 11.84CIGCRR80 pKa = 11.84MEE82 pKa = 5.84KK83 pKa = 10.13ARR85 pKa = 11.84QWGLRR90 pKa = 11.84CLHH93 pKa = 6.31EE94 pKa = 4.94KK95 pKa = 10.49KK96 pKa = 10.39LWAHH100 pKa = 4.84NHH102 pKa = 4.89YY103 pKa = 9.25VTLTYY108 pKa = 10.67NDD110 pKa = 3.6EE111 pKa = 4.12FLPPGGSVCLRR122 pKa = 11.84DD123 pKa = 3.38VQLFMKK129 pKa = 10.4KK130 pKa = 9.49LRR132 pKa = 11.84KK133 pKa = 9.62AKK135 pKa = 10.21GSCKK139 pKa = 9.62EE140 pKa = 3.75NPVRR144 pKa = 11.84FFLGAEE150 pKa = 3.97YY151 pKa = 10.77GDD153 pKa = 3.98EE154 pKa = 4.22NKK156 pKa = 10.21RR157 pKa = 11.84PHH159 pKa = 5.21YY160 pKa = 10.18HH161 pKa = 7.09ALLFNCEE168 pKa = 3.74FNDD171 pKa = 4.32RR172 pKa = 11.84VFWTYY177 pKa = 10.35NKK179 pKa = 10.11RR180 pKa = 11.84GEE182 pKa = 4.1PLYY185 pKa = 10.38TSRR188 pKa = 11.84EE189 pKa = 4.14LTDD192 pKa = 3.03LWGAGHH198 pKa = 7.01CSLGEE203 pKa = 3.94VTFDD207 pKa = 3.85SAVYY211 pKa = 8.51CAKK214 pKa = 10.69YY215 pKa = 10.77AMKK218 pKa = 10.41KK219 pKa = 8.28ITISDD224 pKa = 3.69HH225 pKa = 6.31SSEE228 pKa = 4.19EE229 pKa = 3.75ARR231 pKa = 11.84SEE233 pKa = 3.9FNRR236 pKa = 11.84RR237 pKa = 11.84YY238 pKa = 9.88VVYY241 pKa = 10.36DD242 pKa = 3.19EE243 pKa = 4.41YY244 pKa = 11.9GEE246 pKa = 4.16VFEE249 pKa = 5.28RR250 pKa = 11.84SPEE253 pKa = 3.86FAVMSRR259 pKa = 11.84RR260 pKa = 11.84PGIGAGYY267 pKa = 8.92YY268 pKa = 10.39DD269 pKa = 3.89RR270 pKa = 11.84FGSEE274 pKa = 3.45IRR276 pKa = 11.84THH278 pKa = 7.02DD279 pKa = 3.6SVVVNGRR286 pKa = 11.84EE287 pKa = 3.8VRR289 pKa = 11.84PTRR292 pKa = 11.84FYY294 pKa = 11.11DD295 pKa = 3.58VRR297 pKa = 11.84SEE299 pKa = 4.03RR300 pKa = 11.84CDD302 pKa = 3.39PEE304 pKa = 4.17RR305 pKa = 11.84FAVVKK310 pKa = 10.74AEE312 pKa = 4.54RR313 pKa = 11.84KK314 pKa = 8.32RR315 pKa = 11.84QAVKK319 pKa = 10.88DD320 pKa = 3.79SVKK323 pKa = 10.89ADD325 pKa = 3.48NTPEE329 pKa = 3.89RR330 pKa = 11.84LRR332 pKa = 11.84VKK334 pKa = 10.35EE335 pKa = 4.03VLLEE339 pKa = 4.08AASKK343 pKa = 10.51KK344 pKa = 10.23KK345 pKa = 10.08EE346 pKa = 3.72RR347 pKa = 11.84SLL349 pKa = 3.86
MM1 pKa = 7.9RR2 pKa = 11.84CHH4 pKa = 5.3QAVVSVHH11 pKa = 6.02LVRR14 pKa = 11.84ALVILIRR21 pKa = 11.84RR22 pKa = 11.84IFLLVSQCVAVFGCKK37 pKa = 8.95MGCVKK42 pKa = 10.11PLKK45 pKa = 10.36AFWRR49 pKa = 11.84SRR51 pKa = 11.84SRR53 pKa = 11.84DD54 pKa = 3.5AITFDD59 pKa = 3.35INKK62 pKa = 8.99SATRR66 pKa = 11.84ISFSLPCGRR75 pKa = 11.84CIGCRR80 pKa = 11.84MEE82 pKa = 5.84KK83 pKa = 10.13ARR85 pKa = 11.84QWGLRR90 pKa = 11.84CLHH93 pKa = 6.31EE94 pKa = 4.94KK95 pKa = 10.49KK96 pKa = 10.39LWAHH100 pKa = 4.84NHH102 pKa = 4.89YY103 pKa = 9.25VTLTYY108 pKa = 10.67NDD110 pKa = 3.6EE111 pKa = 4.12FLPPGGSVCLRR122 pKa = 11.84DD123 pKa = 3.38VQLFMKK129 pKa = 10.4KK130 pKa = 9.49LRR132 pKa = 11.84KK133 pKa = 9.62AKK135 pKa = 10.21GSCKK139 pKa = 9.62EE140 pKa = 3.75NPVRR144 pKa = 11.84FFLGAEE150 pKa = 3.97YY151 pKa = 10.77GDD153 pKa = 3.98EE154 pKa = 4.22NKK156 pKa = 10.21RR157 pKa = 11.84PHH159 pKa = 5.21YY160 pKa = 10.18HH161 pKa = 7.09ALLFNCEE168 pKa = 3.74FNDD171 pKa = 4.32RR172 pKa = 11.84VFWTYY177 pKa = 10.35NKK179 pKa = 10.11RR180 pKa = 11.84GEE182 pKa = 4.1PLYY185 pKa = 10.38TSRR188 pKa = 11.84EE189 pKa = 4.14LTDD192 pKa = 3.03LWGAGHH198 pKa = 7.01CSLGEE203 pKa = 3.94VTFDD207 pKa = 3.85SAVYY211 pKa = 8.51CAKK214 pKa = 10.69YY215 pKa = 10.77AMKK218 pKa = 10.41KK219 pKa = 8.28ITISDD224 pKa = 3.69HH225 pKa = 6.31SSEE228 pKa = 4.19EE229 pKa = 3.75ARR231 pKa = 11.84SEE233 pKa = 3.9FNRR236 pKa = 11.84RR237 pKa = 11.84YY238 pKa = 9.88VVYY241 pKa = 10.36DD242 pKa = 3.19EE243 pKa = 4.41YY244 pKa = 11.9GEE246 pKa = 4.16VFEE249 pKa = 5.28RR250 pKa = 11.84SPEE253 pKa = 3.86FAVMSRR259 pKa = 11.84RR260 pKa = 11.84PGIGAGYY267 pKa = 8.92YY268 pKa = 10.39DD269 pKa = 3.89RR270 pKa = 11.84FGSEE274 pKa = 3.45IRR276 pKa = 11.84THH278 pKa = 7.02DD279 pKa = 3.6SVVVNGRR286 pKa = 11.84EE287 pKa = 3.8VRR289 pKa = 11.84PTRR292 pKa = 11.84FYY294 pKa = 11.11DD295 pKa = 3.58VRR297 pKa = 11.84SEE299 pKa = 4.03RR300 pKa = 11.84CDD302 pKa = 3.39PEE304 pKa = 4.17RR305 pKa = 11.84FAVVKK310 pKa = 10.74AEE312 pKa = 4.54RR313 pKa = 11.84KK314 pKa = 8.32RR315 pKa = 11.84QAVKK319 pKa = 10.88DD320 pKa = 3.79SVKK323 pKa = 10.89ADD325 pKa = 3.48NTPEE329 pKa = 3.89RR330 pKa = 11.84LRR332 pKa = 11.84VKK334 pKa = 10.35EE335 pKa = 4.03VLLEE339 pKa = 4.08AASKK343 pKa = 10.51KK344 pKa = 10.23KK345 pKa = 10.08EE346 pKa = 3.72RR347 pKa = 11.84SLL349 pKa = 3.86
Molecular weight: 40.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1422 |
90 |
553 |
284.4 |
31.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.861 ± 1.081 | 1.406 ± 0.76 |
4.852 ± 0.369 | 5.415 ± 1.601 |
5.696 ± 0.984 | 7.032 ± 0.491 |
1.758 ± 0.463 | 3.376 ± 0.436 |
4.36 ± 1.244 | 7.173 ± 0.493 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.672 ± 0.659 | 4.219 ± 0.448 |
6.118 ± 1.21 | 4.29 ± 0.982 |
7.173 ± 1.073 | 8.579 ± 0.977 |
6.399 ± 1.287 | 6.048 ± 0.993 |
1.266 ± 0.128 | 3.305 ± 0.772 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
