
Itaporanga virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Phenuiviridae; Phlebovirus; Itaporanga phlebovirus
Average proteome isoelectric point is 6.46
Get precalculated fractions of proteins
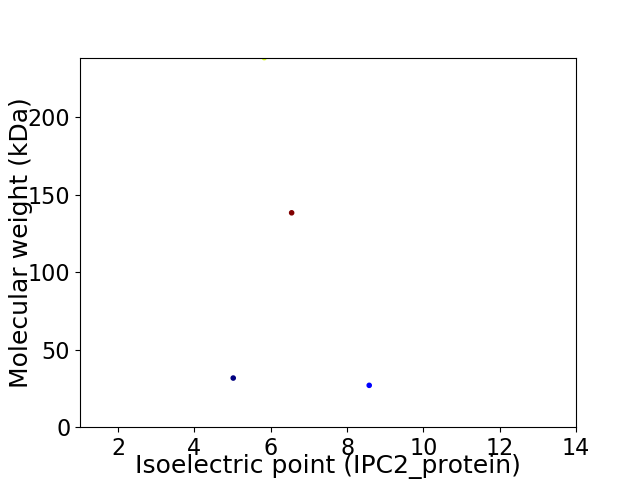
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4V1DW07|A0A4V1DW07_9VIRU Replicase OS=Itaporanga virus OX=629735 PE=4 SV=2
MM1 pKa = 7.11MRR3 pKa = 11.84FFHH6 pKa = 7.32PEE8 pKa = 3.38MPEE11 pKa = 3.64YY12 pKa = 11.01SFDD15 pKa = 3.6YY16 pKa = 10.1SKK18 pKa = 11.42EE19 pKa = 3.86GGRR22 pKa = 11.84VNVLYY27 pKa = 10.86VNKK30 pKa = 10.13FEE32 pKa = 4.22WTEE35 pKa = 4.22VPASYY40 pKa = 11.02HH41 pKa = 5.66MGSLIPLKK49 pKa = 10.31FVSLSRR55 pKa = 11.84KK56 pKa = 9.76KK57 pKa = 10.74FMNLKK62 pKa = 9.85EE63 pKa = 4.2FYY65 pKa = 10.78DD66 pKa = 4.5RR67 pKa = 11.84DD68 pKa = 4.11TLPLLWGDD76 pKa = 3.99CKK78 pKa = 10.76GSQVMGPSLNCFDD91 pKa = 5.47EE92 pKa = 4.68MIKK95 pKa = 9.62MLASIPLNDD104 pKa = 4.03FLAPSRR110 pKa = 11.84GHH112 pKa = 5.29TAHH115 pKa = 7.17GISWPTGSPDD125 pKa = 3.33LNYY128 pKa = 9.97IVYY131 pKa = 10.02SSKK134 pKa = 10.96DD135 pKa = 3.51INPEE139 pKa = 3.83SYY141 pKa = 10.69YY142 pKa = 10.74SRR144 pKa = 11.84CHH146 pKa = 5.0VASMLMNGAKK156 pKa = 10.31GNPSLDD162 pKa = 3.83LACVALHH169 pKa = 5.93KK170 pKa = 10.32KK171 pKa = 8.69IRR173 pKa = 11.84KK174 pKa = 8.42AAADD178 pKa = 3.42MGVSPEE184 pKa = 4.18LFSGRR189 pKa = 11.84NLVKK193 pKa = 10.56DD194 pKa = 3.78VACVQVISVLRR205 pKa = 11.84GAEE208 pKa = 3.43MDD210 pKa = 3.19QRR212 pKa = 11.84VFGKK216 pKa = 10.64GDD218 pKa = 3.65NLVDD222 pKa = 3.56WLVEE226 pKa = 3.67CAARR230 pKa = 11.84IRR232 pKa = 11.84TSDD235 pKa = 3.64PGCLGDD241 pKa = 4.86EE242 pKa = 4.1IGFGPRR248 pKa = 11.84CRR250 pKa = 11.84AIDD253 pKa = 3.98DD254 pKa = 4.3LDD256 pKa = 3.9RR257 pKa = 11.84CLEE260 pKa = 4.33GNDD263 pKa = 4.2SDD265 pKa = 5.76FSDD268 pKa = 5.13DD269 pKa = 4.51SSDD272 pKa = 4.1DD273 pKa = 3.42EE274 pKa = 4.46AAIAASNLSSVEE286 pKa = 3.85
MM1 pKa = 7.11MRR3 pKa = 11.84FFHH6 pKa = 7.32PEE8 pKa = 3.38MPEE11 pKa = 3.64YY12 pKa = 11.01SFDD15 pKa = 3.6YY16 pKa = 10.1SKK18 pKa = 11.42EE19 pKa = 3.86GGRR22 pKa = 11.84VNVLYY27 pKa = 10.86VNKK30 pKa = 10.13FEE32 pKa = 4.22WTEE35 pKa = 4.22VPASYY40 pKa = 11.02HH41 pKa = 5.66MGSLIPLKK49 pKa = 10.31FVSLSRR55 pKa = 11.84KK56 pKa = 9.76KK57 pKa = 10.74FMNLKK62 pKa = 9.85EE63 pKa = 4.2FYY65 pKa = 10.78DD66 pKa = 4.5RR67 pKa = 11.84DD68 pKa = 4.11TLPLLWGDD76 pKa = 3.99CKK78 pKa = 10.76GSQVMGPSLNCFDD91 pKa = 5.47EE92 pKa = 4.68MIKK95 pKa = 9.62MLASIPLNDD104 pKa = 4.03FLAPSRR110 pKa = 11.84GHH112 pKa = 5.29TAHH115 pKa = 7.17GISWPTGSPDD125 pKa = 3.33LNYY128 pKa = 9.97IVYY131 pKa = 10.02SSKK134 pKa = 10.96DD135 pKa = 3.51INPEE139 pKa = 3.83SYY141 pKa = 10.69YY142 pKa = 10.74SRR144 pKa = 11.84CHH146 pKa = 5.0VASMLMNGAKK156 pKa = 10.31GNPSLDD162 pKa = 3.83LACVALHH169 pKa = 5.93KK170 pKa = 10.32KK171 pKa = 8.69IRR173 pKa = 11.84KK174 pKa = 8.42AAADD178 pKa = 3.42MGVSPEE184 pKa = 4.18LFSGRR189 pKa = 11.84NLVKK193 pKa = 10.56DD194 pKa = 3.78VACVQVISVLRR205 pKa = 11.84GAEE208 pKa = 3.43MDD210 pKa = 3.19QRR212 pKa = 11.84VFGKK216 pKa = 10.64GDD218 pKa = 3.65NLVDD222 pKa = 3.56WLVEE226 pKa = 3.67CAARR230 pKa = 11.84IRR232 pKa = 11.84TSDD235 pKa = 3.64PGCLGDD241 pKa = 4.86EE242 pKa = 4.1IGFGPRR248 pKa = 11.84CRR250 pKa = 11.84AIDD253 pKa = 3.98DD254 pKa = 4.3LDD256 pKa = 3.9RR257 pKa = 11.84CLEE260 pKa = 4.33GNDD263 pKa = 4.2SDD265 pKa = 5.76FSDD268 pKa = 5.13DD269 pKa = 4.51SSDD272 pKa = 4.1DD273 pKa = 3.42EE274 pKa = 4.46AAIAASNLSSVEE286 pKa = 3.85
Molecular weight: 31.71 kDa
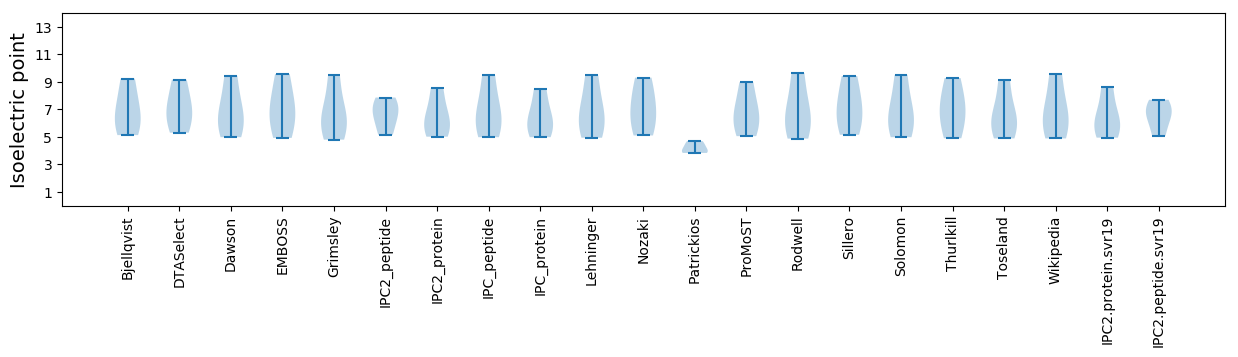
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8D7T3|A0A4P8D7T3_9VIRU Glycoprotein OS=Itaporanga virus OX=629735 PE=4 SV=1
MM1 pKa = 7.65SDD3 pKa = 3.5YY4 pKa = 11.5ARR6 pKa = 11.84IALDD10 pKa = 4.11FYY12 pKa = 11.25EE13 pKa = 4.79NNPPTTNDD21 pKa = 2.65IAEE24 pKa = 4.28FVSLFAYY31 pKa = 9.81QGYY34 pKa = 9.1DD35 pKa = 2.58ARR37 pKa = 11.84TVVRR41 pKa = 11.84LVTEE45 pKa = 4.32RR46 pKa = 11.84GGGNWQEE53 pKa = 3.92DD54 pKa = 4.1VKK56 pKa = 11.21RR57 pKa = 11.84LIVLAATRR65 pKa = 11.84GNRR68 pKa = 11.84VNKK71 pKa = 9.29MKK73 pKa = 11.02AKK75 pKa = 10.07MSDD78 pKa = 3.05KK79 pKa = 11.15GKK81 pKa = 11.3GEE83 pKa = 3.96LTKK86 pKa = 10.83LVSTYY91 pKa = 10.65KK92 pKa = 10.61LKK94 pKa = 10.77EE95 pKa = 4.07GNPGRR100 pKa = 11.84EE101 pKa = 4.3DD102 pKa = 3.24LTLSRR107 pKa = 11.84VACAFATWTCTCITSIAEE125 pKa = 4.02YY126 pKa = 10.71LPVTGSTMDD135 pKa = 3.51SYY137 pKa = 11.8SPNYY141 pKa = 8.48PRR143 pKa = 11.84TMMHH147 pKa = 6.89PAFGGLIDD155 pKa = 3.99LTLPEE160 pKa = 5.24AEE162 pKa = 4.39TLVAAHH168 pKa = 6.56SLYY171 pKa = 10.94LNRR174 pKa = 11.84FAQVINPGLRR184 pKa = 11.84GKK186 pKa = 10.57SKK188 pKa = 11.36DD189 pKa = 3.51EE190 pKa = 3.86IQATFQQPLNAAANSSFITSEE211 pKa = 3.95NKK213 pKa = 9.94RR214 pKa = 11.84KK215 pKa = 9.52FLTKK219 pKa = 10.42FMLIDD224 pKa = 3.88SNLKK228 pKa = 7.46VTKK231 pKa = 10.36SVADD235 pKa = 3.35AAKK238 pKa = 10.18AYY240 pKa = 9.06KK241 pKa = 10.33ALNN244 pKa = 3.6
MM1 pKa = 7.65SDD3 pKa = 3.5YY4 pKa = 11.5ARR6 pKa = 11.84IALDD10 pKa = 4.11FYY12 pKa = 11.25EE13 pKa = 4.79NNPPTTNDD21 pKa = 2.65IAEE24 pKa = 4.28FVSLFAYY31 pKa = 9.81QGYY34 pKa = 9.1DD35 pKa = 2.58ARR37 pKa = 11.84TVVRR41 pKa = 11.84LVTEE45 pKa = 4.32RR46 pKa = 11.84GGGNWQEE53 pKa = 3.92DD54 pKa = 4.1VKK56 pKa = 11.21RR57 pKa = 11.84LIVLAATRR65 pKa = 11.84GNRR68 pKa = 11.84VNKK71 pKa = 9.29MKK73 pKa = 11.02AKK75 pKa = 10.07MSDD78 pKa = 3.05KK79 pKa = 11.15GKK81 pKa = 11.3GEE83 pKa = 3.96LTKK86 pKa = 10.83LVSTYY91 pKa = 10.65KK92 pKa = 10.61LKK94 pKa = 10.77EE95 pKa = 4.07GNPGRR100 pKa = 11.84EE101 pKa = 4.3DD102 pKa = 3.24LTLSRR107 pKa = 11.84VACAFATWTCTCITSIAEE125 pKa = 4.02YY126 pKa = 10.71LPVTGSTMDD135 pKa = 3.51SYY137 pKa = 11.8SPNYY141 pKa = 8.48PRR143 pKa = 11.84TMMHH147 pKa = 6.89PAFGGLIDD155 pKa = 3.99LTLPEE160 pKa = 5.24AEE162 pKa = 4.39TLVAAHH168 pKa = 6.56SLYY171 pKa = 10.94LNRR174 pKa = 11.84FAQVINPGLRR184 pKa = 11.84GKK186 pKa = 10.57SKK188 pKa = 11.36DD189 pKa = 3.51EE190 pKa = 3.86IQATFQQPLNAAANSSFITSEE211 pKa = 3.95NKK213 pKa = 9.94RR214 pKa = 11.84KK215 pKa = 9.52FLTKK219 pKa = 10.42FMLIDD224 pKa = 3.88SNLKK228 pKa = 7.46VTKK231 pKa = 10.36SVADD235 pKa = 3.35AAKK238 pKa = 10.18AYY240 pKa = 9.06KK241 pKa = 10.33ALNN244 pKa = 3.6
Molecular weight: 27.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3890 |
244 |
2094 |
972.5 |
108.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.578 ± 0.987 | 2.802 ± 0.876 |
5.604 ± 0.564 | 6.607 ± 0.807 |
4.576 ± 0.121 | 6.375 ± 0.442 |
2.262 ± 0.274 | 6.298 ± 0.603 |
6.35 ± 0.42 | 8.612 ± 0.249 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.828 ± 0.649 | 3.702 ± 0.491 |
4.293 ± 0.363 | 2.931 ± 0.444 |
5.193 ± 0.392 | 9.486 ± 1.009 |
5.681 ± 0.584 | 6.735 ± 0.273 |
1.183 ± 0.089 | 2.905 ± 0.295 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
