
Penguinpox virus
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Nucleocytoviricota; Pokkesviricetes; Chitovirales; Poxviridae; Chordopoxvirinae; Avipoxvirus
Average proteome isoelectric point is 6.89
Get precalculated fractions of proteins
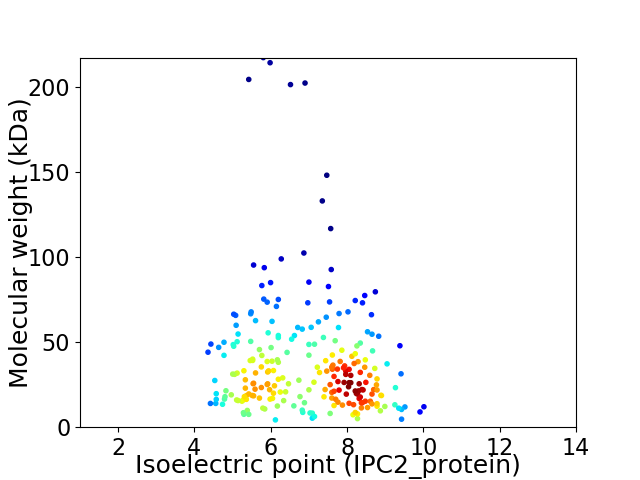
Virtual 2D-PAGE plot for 240 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
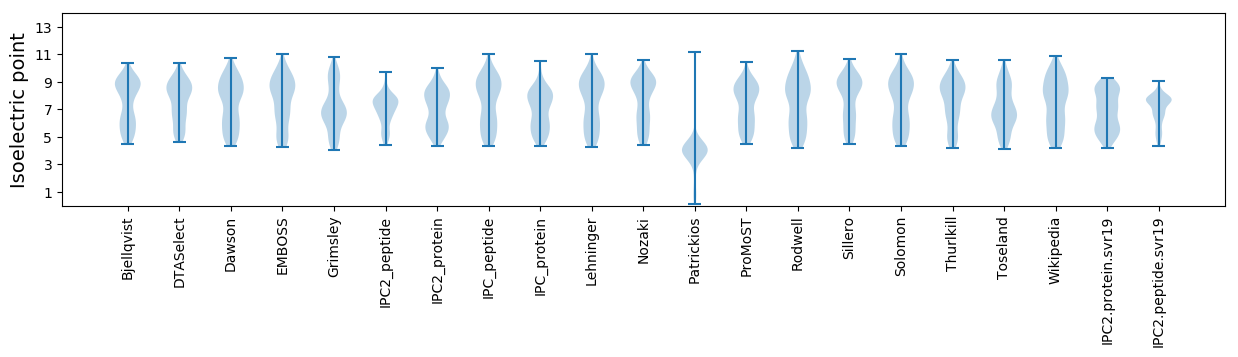
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A068ELR0|A0A068ELR0_9POXV DNA helicase OS=Penguinpox virus OX=648998 GN=pepv_196 PE=4 SV=1
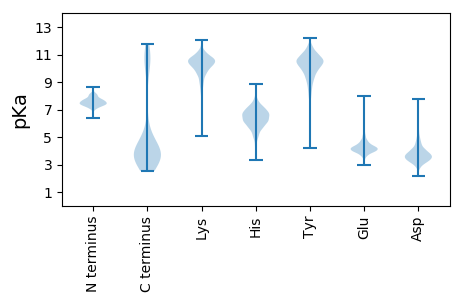
MM1 pKa = 7.38QYY3 pKa = 9.66TRR5 pKa = 11.84IIMIYY10 pKa = 9.77IITSLLFIKK19 pKa = 9.3VTTGITEE26 pKa = 4.24PVKK29 pKa = 10.44NPQDD33 pKa = 3.17ILHH36 pKa = 6.3IMEE39 pKa = 4.68HH40 pKa = 6.13NKK42 pKa = 9.63TGVTACSLYY51 pKa = 11.04CFDD54 pKa = 4.41QSKK57 pKa = 10.58GLDD60 pKa = 3.39QPKK63 pKa = 8.87TFILPGKK70 pKa = 9.66YY71 pKa = 9.99SNNSIKK77 pKa = 10.84LEE79 pKa = 3.9IAIDD83 pKa = 3.65IYY85 pKa = 11.5KK86 pKa = 10.12KK87 pKa = 10.36DD88 pKa = 3.62SKK90 pKa = 10.72PDD92 pKa = 3.47YY93 pKa = 10.52SHH95 pKa = 7.76PCQAFQFCVSGNFSGKK111 pKa = 10.17RR112 pKa = 11.84FDD114 pKa = 3.75HH115 pKa = 6.1YY116 pKa = 11.25LYY118 pKa = 10.52GYY120 pKa = 7.99TITGFIDD127 pKa = 3.19IASSYY132 pKa = 9.19YY133 pKa = 10.61SGMSISTITLMPLQEE148 pKa = 4.61GSLNPDD154 pKa = 3.25SEE156 pKa = 5.15DD157 pKa = 4.43EE158 pKa = 4.33EE159 pKa = 6.24DD160 pKa = 4.12CTTPPISTITQSQRR174 pKa = 11.84IPEE177 pKa = 4.04PVIKK181 pKa = 10.04EE182 pKa = 3.72GCRR185 pKa = 11.84PVVLQRR191 pKa = 11.84YY192 pKa = 9.17AEE194 pKa = 4.38SDD196 pKa = 3.75DD197 pKa = 4.11PTCIMYY203 pKa = 9.41WDD205 pKa = 4.17HH206 pKa = 6.67TWDD209 pKa = 3.55NYY211 pKa = 10.85CDD213 pKa = 3.29VGFFNSQQRR222 pKa = 11.84DD223 pKa = 3.31HH224 pKa = 7.54DD225 pKa = 4.28PLVFPLKK232 pKa = 10.32SYY234 pKa = 10.87LGIRR238 pKa = 11.84GAFQDD243 pKa = 3.92FQSYY247 pKa = 8.5YY248 pKa = 10.68CKK250 pKa = 10.68SLDD253 pKa = 3.71LNQSYY258 pKa = 9.43SVCISIGEE266 pKa = 4.15TPIAVTYY273 pKa = 10.26HH274 pKa = 6.11SYY276 pKa = 11.73EE277 pKa = 4.28NITVNEE283 pKa = 4.11LLTRR287 pKa = 11.84IMTLYY292 pKa = 10.96GEE294 pKa = 4.58EE295 pKa = 4.21NVHH298 pKa = 6.3KK299 pKa = 10.57LPYY302 pKa = 10.14RR303 pKa = 11.84NITIMAHH310 pKa = 5.56AQIQSLPLINGTCDD324 pKa = 3.3TDD326 pKa = 3.88GSNDD330 pKa = 3.89DD331 pKa = 4.61SDD333 pKa = 5.77SGDD336 pKa = 5.18DD337 pKa = 5.89DD338 pKa = 6.88DD339 pKa = 7.74DD340 pKa = 7.12DD341 pKa = 6.72DD342 pKa = 6.75DD343 pKa = 6.4YY344 pKa = 12.06YY345 pKa = 11.56DD346 pKa = 4.8EE347 pKa = 4.85YY348 pKa = 11.75DD349 pKa = 4.31LYY351 pKa = 10.77IEE353 pKa = 4.29STPSIVTVKK362 pKa = 10.2PKK364 pKa = 9.65KK365 pKa = 9.08TVTDD369 pKa = 4.08EE370 pKa = 4.06YY371 pKa = 11.72NSIFNSFDD379 pKa = 3.22NFDD382 pKa = 3.66LEE384 pKa = 4.61KK385 pKa = 10.78RR386 pKa = 3.75
MM1 pKa = 7.38QYY3 pKa = 9.66TRR5 pKa = 11.84IIMIYY10 pKa = 9.77IITSLLFIKK19 pKa = 9.3VTTGITEE26 pKa = 4.24PVKK29 pKa = 10.44NPQDD33 pKa = 3.17ILHH36 pKa = 6.3IMEE39 pKa = 4.68HH40 pKa = 6.13NKK42 pKa = 9.63TGVTACSLYY51 pKa = 11.04CFDD54 pKa = 4.41QSKK57 pKa = 10.58GLDD60 pKa = 3.39QPKK63 pKa = 8.87TFILPGKK70 pKa = 9.66YY71 pKa = 9.99SNNSIKK77 pKa = 10.84LEE79 pKa = 3.9IAIDD83 pKa = 3.65IYY85 pKa = 11.5KK86 pKa = 10.12KK87 pKa = 10.36DD88 pKa = 3.62SKK90 pKa = 10.72PDD92 pKa = 3.47YY93 pKa = 10.52SHH95 pKa = 7.76PCQAFQFCVSGNFSGKK111 pKa = 10.17RR112 pKa = 11.84FDD114 pKa = 3.75HH115 pKa = 6.1YY116 pKa = 11.25LYY118 pKa = 10.52GYY120 pKa = 7.99TITGFIDD127 pKa = 3.19IASSYY132 pKa = 9.19YY133 pKa = 10.61SGMSISTITLMPLQEE148 pKa = 4.61GSLNPDD154 pKa = 3.25SEE156 pKa = 5.15DD157 pKa = 4.43EE158 pKa = 4.33EE159 pKa = 6.24DD160 pKa = 4.12CTTPPISTITQSQRR174 pKa = 11.84IPEE177 pKa = 4.04PVIKK181 pKa = 10.04EE182 pKa = 3.72GCRR185 pKa = 11.84PVVLQRR191 pKa = 11.84YY192 pKa = 9.17AEE194 pKa = 4.38SDD196 pKa = 3.75DD197 pKa = 4.11PTCIMYY203 pKa = 9.41WDD205 pKa = 4.17HH206 pKa = 6.67TWDD209 pKa = 3.55NYY211 pKa = 10.85CDD213 pKa = 3.29VGFFNSQQRR222 pKa = 11.84DD223 pKa = 3.31HH224 pKa = 7.54DD225 pKa = 4.28PLVFPLKK232 pKa = 10.32SYY234 pKa = 10.87LGIRR238 pKa = 11.84GAFQDD243 pKa = 3.92FQSYY247 pKa = 8.5YY248 pKa = 10.68CKK250 pKa = 10.68SLDD253 pKa = 3.71LNQSYY258 pKa = 9.43SVCISIGEE266 pKa = 4.15TPIAVTYY273 pKa = 10.26HH274 pKa = 6.11SYY276 pKa = 11.73EE277 pKa = 4.28NITVNEE283 pKa = 4.11LLTRR287 pKa = 11.84IMTLYY292 pKa = 10.96GEE294 pKa = 4.58EE295 pKa = 4.21NVHH298 pKa = 6.3KK299 pKa = 10.57LPYY302 pKa = 10.14RR303 pKa = 11.84NITIMAHH310 pKa = 5.56AQIQSLPLINGTCDD324 pKa = 3.3TDD326 pKa = 3.88GSNDD330 pKa = 3.89DD331 pKa = 4.61SDD333 pKa = 5.77SGDD336 pKa = 5.18DD337 pKa = 5.89DD338 pKa = 6.88DD339 pKa = 7.74DD340 pKa = 7.12DD341 pKa = 6.72DD342 pKa = 6.75DD343 pKa = 6.4YY344 pKa = 12.06YY345 pKa = 11.56DD346 pKa = 4.8EE347 pKa = 4.85YY348 pKa = 11.75DD349 pKa = 4.31LYY351 pKa = 10.77IEE353 pKa = 4.29STPSIVTVKK362 pKa = 10.2PKK364 pKa = 9.65KK365 pKa = 9.08TVTDD369 pKa = 4.08EE370 pKa = 4.06YY371 pKa = 11.72NSIFNSFDD379 pKa = 3.22NFDD382 pKa = 3.66LEE384 pKa = 4.61KK385 pKa = 10.78RR386 pKa = 3.75
Molecular weight: 44.07 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A068EHK9|A0A068EHK9_9POXV Telomere-binding protein I1 OS=Penguinpox virus OX=648998 GN=pepv_093 PE=3 SV=1
MM1 pKa = 7.54EE2 pKa = 5.37IARR5 pKa = 11.84EE6 pKa = 3.92TLITIGLTILVVVLVITGFSLVLRR30 pKa = 11.84LIPGVYY36 pKa = 9.71SAASRR41 pKa = 11.84SSFTAGKK48 pKa = 9.17VLRR51 pKa = 11.84FMEE54 pKa = 4.3IFSTVMFIPGIIILYY69 pKa = 9.17AAYY72 pKa = 9.46IRR74 pKa = 11.84KK75 pKa = 7.84TKK77 pKa = 9.57MKK79 pKa = 10.73NNN81 pKa = 3.69
MM1 pKa = 7.54EE2 pKa = 5.37IARR5 pKa = 11.84EE6 pKa = 3.92TLITIGLTILVVVLVITGFSLVLRR30 pKa = 11.84LIPGVYY36 pKa = 9.71SAASRR41 pKa = 11.84SSFTAGKK48 pKa = 9.17VLRR51 pKa = 11.84FMEE54 pKa = 4.3IFSTVMFIPGIIILYY69 pKa = 9.17AAYY72 pKa = 9.46IRR74 pKa = 11.84KK75 pKa = 7.84TKK77 pKa = 9.57MKK79 pKa = 10.73NNN81 pKa = 3.69
Molecular weight: 8.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
80106 |
34 |
1922 |
333.8 |
38.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.423 ± 0.104 | 2.177 ± 0.087 |
6.33 ± 0.114 | 5.68 ± 0.09 |
4.197 ± 0.109 | 3.902 ± 0.107 |
2.016 ± 0.043 | 10.281 ± 0.175 |
8.359 ± 0.151 | 9.2 ± 0.194 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.552 ± 0.06 | 7.473 ± 0.16 |
3.081 ± 0.106 | 1.909 ± 0.059 |
3.967 ± 0.1 | 7.758 ± 0.201 |
5.634 ± 0.116 | 5.661 ± 0.092 |
0.643 ± 0.043 | 5.756 ± 0.089 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
