
Aliikangiella marina
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Oceanospirillales; Pleioneaceae; Aliikangiella
Average proteome isoelectric point is 6.18
Get precalculated fractions of proteins
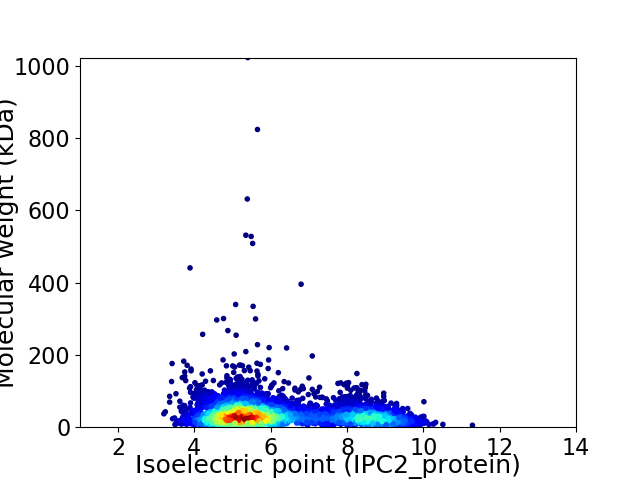
Virtual 2D-PAGE plot for 4367 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A545T2U9|A0A545T2U9_9GAMM Maleylacetoacetate isomerase OS=Aliikangiella marina OX=1712262 GN=maiA PE=3 SV=1
MM1 pKa = 7.59INLKK5 pKa = 10.47KK6 pKa = 10.64SLLAGAIAACSLSAAAVEE24 pKa = 4.1TAYY27 pKa = 11.14NIVPLGTFEE36 pKa = 5.12DD37 pKa = 3.99NEE39 pKa = 3.96IFAFGINNNDD49 pKa = 3.1VAVGYY54 pKa = 10.44SYY56 pKa = 10.49VTGGFPSRR64 pKa = 11.84GYY66 pKa = 11.03AYY68 pKa = 10.58DD69 pKa = 3.88NGTFTNMGSIVNQEE83 pKa = 3.83LAIGFIAATIDD94 pKa = 3.24EE95 pKa = 4.76SVAFDD100 pKa = 3.57INDD103 pKa = 3.34NGMMIGYY110 pKa = 9.0SVEE113 pKa = 4.2SVPQFDD119 pKa = 4.53EE120 pKa = 4.65NNDD123 pKa = 3.75PVTVTVDD130 pKa = 3.03QGLDD134 pKa = 3.41TEE136 pKa = 5.33RR137 pKa = 11.84EE138 pKa = 4.26LQSGLPVQRR147 pKa = 11.84AIYY150 pKa = 8.01TQVGSQVLSQVPLFEE165 pKa = 5.07PEE167 pKa = 3.38QDD169 pKa = 3.1HH170 pKa = 6.26GVRR173 pKa = 11.84ALAVNNNGVIVGFAEE188 pKa = 5.04LDD190 pKa = 3.46VDD192 pKa = 5.49DD193 pKa = 6.0DD194 pKa = 4.08LTAEE198 pKa = 4.45GEE200 pKa = 4.08DD201 pKa = 3.79AEE203 pKa = 5.03FIVNRR208 pKa = 11.84GVIYY212 pKa = 8.03DD213 pKa = 3.99TQTDD217 pKa = 3.54TLTRR221 pKa = 11.84VNPLNYY227 pKa = 10.25DD228 pKa = 3.49NLPRR232 pKa = 11.84SIALKK237 pKa = 10.27DD238 pKa = 3.37INDD241 pKa = 3.57NGVAVGWSQVAFEE254 pKa = 5.07SSFLFNGVVVNAASPEE270 pKa = 4.18TLTTIDD276 pKa = 4.2PGEE279 pKa = 4.12EE280 pKa = 3.7TASLFNAINNNGMVAGKK297 pKa = 10.45LYY299 pKa = 10.77DD300 pKa = 3.76LEE302 pKa = 4.15KK303 pKa = 10.53QRR305 pKa = 11.84YY306 pKa = 7.19EE307 pKa = 4.15GIMFNSNNGAFEE319 pKa = 4.91IIPPLVEE326 pKa = 4.79GLDD329 pKa = 3.96SQICCDD335 pKa = 3.47NASPYY340 pKa = 10.49DD341 pKa = 3.67INAQNIIVGSMIADD355 pKa = 3.78VVPDD359 pKa = 4.1TYY361 pKa = 11.16HH362 pKa = 7.16AFAYY366 pKa = 10.45QNGTTYY372 pKa = 11.34DD373 pKa = 3.92LNDD376 pKa = 5.88LIDD379 pKa = 4.07CTQTLTTGVRR389 pKa = 11.84DD390 pKa = 3.41WVLYY394 pKa = 7.6EE395 pKa = 4.07ARR397 pKa = 11.84QINDD401 pKa = 2.89NGIIIGNGLFGGEE414 pKa = 3.84RR415 pKa = 11.84KK416 pKa = 10.33AFMLTPNPGATPTQCDD432 pKa = 3.92FEE434 pKa = 4.61QEE436 pKa = 4.35EE437 pKa = 4.52EE438 pKa = 4.28DD439 pKa = 4.19SGSGSIPASLIVLLSCLAVIRR460 pKa = 11.84RR461 pKa = 11.84KK462 pKa = 10.41VSS464 pKa = 2.84
MM1 pKa = 7.59INLKK5 pKa = 10.47KK6 pKa = 10.64SLLAGAIAACSLSAAAVEE24 pKa = 4.1TAYY27 pKa = 11.14NIVPLGTFEE36 pKa = 5.12DD37 pKa = 3.99NEE39 pKa = 3.96IFAFGINNNDD49 pKa = 3.1VAVGYY54 pKa = 10.44SYY56 pKa = 10.49VTGGFPSRR64 pKa = 11.84GYY66 pKa = 11.03AYY68 pKa = 10.58DD69 pKa = 3.88NGTFTNMGSIVNQEE83 pKa = 3.83LAIGFIAATIDD94 pKa = 3.24EE95 pKa = 4.76SVAFDD100 pKa = 3.57INDD103 pKa = 3.34NGMMIGYY110 pKa = 9.0SVEE113 pKa = 4.2SVPQFDD119 pKa = 4.53EE120 pKa = 4.65NNDD123 pKa = 3.75PVTVTVDD130 pKa = 3.03QGLDD134 pKa = 3.41TEE136 pKa = 5.33RR137 pKa = 11.84EE138 pKa = 4.26LQSGLPVQRR147 pKa = 11.84AIYY150 pKa = 8.01TQVGSQVLSQVPLFEE165 pKa = 5.07PEE167 pKa = 3.38QDD169 pKa = 3.1HH170 pKa = 6.26GVRR173 pKa = 11.84ALAVNNNGVIVGFAEE188 pKa = 5.04LDD190 pKa = 3.46VDD192 pKa = 5.49DD193 pKa = 6.0DD194 pKa = 4.08LTAEE198 pKa = 4.45GEE200 pKa = 4.08DD201 pKa = 3.79AEE203 pKa = 5.03FIVNRR208 pKa = 11.84GVIYY212 pKa = 8.03DD213 pKa = 3.99TQTDD217 pKa = 3.54TLTRR221 pKa = 11.84VNPLNYY227 pKa = 10.25DD228 pKa = 3.49NLPRR232 pKa = 11.84SIALKK237 pKa = 10.27DD238 pKa = 3.37INDD241 pKa = 3.57NGVAVGWSQVAFEE254 pKa = 5.07SSFLFNGVVVNAASPEE270 pKa = 4.18TLTTIDD276 pKa = 4.2PGEE279 pKa = 4.12EE280 pKa = 3.7TASLFNAINNNGMVAGKK297 pKa = 10.45LYY299 pKa = 10.77DD300 pKa = 3.76LEE302 pKa = 4.15KK303 pKa = 10.53QRR305 pKa = 11.84YY306 pKa = 7.19EE307 pKa = 4.15GIMFNSNNGAFEE319 pKa = 4.91IIPPLVEE326 pKa = 4.79GLDD329 pKa = 3.96SQICCDD335 pKa = 3.47NASPYY340 pKa = 10.49DD341 pKa = 3.67INAQNIIVGSMIADD355 pKa = 3.78VVPDD359 pKa = 4.1TYY361 pKa = 11.16HH362 pKa = 7.16AFAYY366 pKa = 10.45QNGTTYY372 pKa = 11.34DD373 pKa = 3.92LNDD376 pKa = 5.88LIDD379 pKa = 4.07CTQTLTTGVRR389 pKa = 11.84DD390 pKa = 3.41WVLYY394 pKa = 7.6EE395 pKa = 4.07ARR397 pKa = 11.84QINDD401 pKa = 2.89NGIIIGNGLFGGEE414 pKa = 3.84RR415 pKa = 11.84KK416 pKa = 10.33AFMLTPNPGATPTQCDD432 pKa = 3.92FEE434 pKa = 4.61QEE436 pKa = 4.35EE437 pKa = 4.52EE438 pKa = 4.28DD439 pKa = 4.19SGSGSIPASLIVLLSCLAVIRR460 pKa = 11.84RR461 pKa = 11.84KK462 pKa = 10.41VSS464 pKa = 2.84
Molecular weight: 49.96 kDa
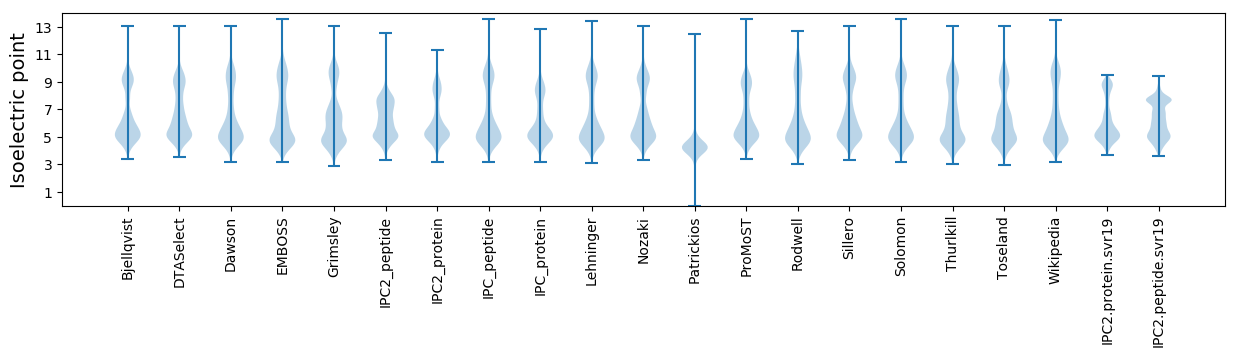
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A545T2F4|A0A545T2F4_9GAMM Branched-chain alpha-keto acid dehydrogenase subunit E2 OS=Aliikangiella marina OX=1712262 GN=FLL45_19770 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSTLKK11 pKa = 10.3RR12 pKa = 11.84KK13 pKa = 9.12RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.47GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.45NGRR28 pKa = 11.84QVINRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.7GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSTLKK11 pKa = 10.3RR12 pKa = 11.84KK13 pKa = 9.12RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.47GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.45NGRR28 pKa = 11.84QVINRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.7GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1527963 |
37 |
8991 |
349.9 |
39.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.959 ± 0.049 | 0.93 ± 0.012 |
5.699 ± 0.03 | 6.593 ± 0.036 |
4.464 ± 0.026 | 6.411 ± 0.037 |
1.987 ± 0.016 | 6.808 ± 0.03 |
5.853 ± 0.038 | 10.073 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.256 ± 0.019 | 4.866 ± 0.03 |
3.798 ± 0.022 | 4.504 ± 0.027 |
4.502 ± 0.025 | 7.139 ± 0.034 |
5.172 ± 0.029 | 6.629 ± 0.028 |
1.244 ± 0.013 | 3.112 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
