
Psittacid herpesvirus 1 (isolate Amazon parrot/-/97-0001/1997) (PsHV-1) (Pacheco s disease virus)
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Herpesviridae; Alphaherpesvirinae; Iltovirus; Psittacid alphaherpesvirus 1
Average proteome isoelectric point is 6.59
Get precalculated fractions of proteins
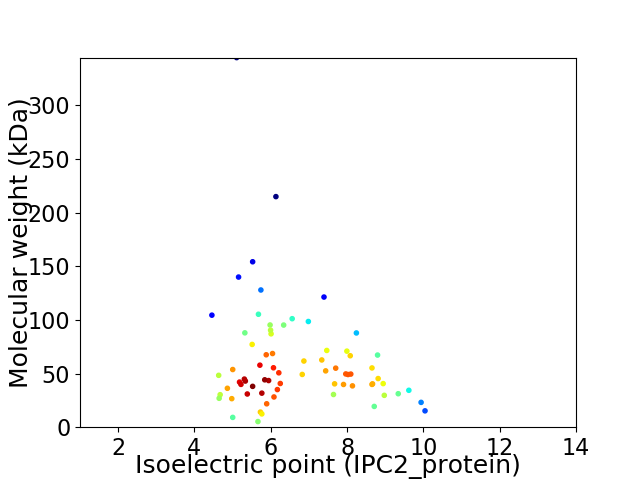
Virtual 2D-PAGE plot for 72 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q6UDF8|GG_PSHV1 Glycoprotein G OS=Psittacid herpesvirus 1 (isolate Amazon parrot/-/97-0001/1997) OX=670426 GN=gG PE=3 SV=1
MM1 pKa = 7.71LWPAALVAMFALAARR16 pKa = 11.84ARR18 pKa = 11.84EE19 pKa = 4.04IDD21 pKa = 3.64NVTCSVVYY29 pKa = 9.66GSNVARR35 pKa = 11.84ISKK38 pKa = 9.95DD39 pKa = 2.97YY40 pKa = 10.5WLAQEE45 pKa = 4.73GSLVSFMTFEE55 pKa = 4.43NDD57 pKa = 3.13DD58 pKa = 3.47PVYY61 pKa = 10.82FFMGRR66 pKa = 11.84ALGTGASGGKK76 pKa = 9.21YY77 pKa = 9.89LYY79 pKa = 10.49RR80 pKa = 11.84VTRR83 pKa = 11.84DD84 pKa = 2.89NNTNVSHH91 pKa = 5.86VQTSLFIPEE100 pKa = 4.62HH101 pKa = 5.8FNGGILLDD109 pKa = 3.48MGSNYY114 pKa = 8.76THH116 pKa = 7.6DD117 pKa = 4.73AEE119 pKa = 4.61TDD121 pKa = 3.68PEE123 pKa = 4.27FLNARR128 pKa = 11.84YY129 pKa = 9.16LVVNDD134 pKa = 4.1TEE136 pKa = 4.73DD137 pKa = 3.35SVRR140 pKa = 11.84EE141 pKa = 4.05EE142 pKa = 4.17TLGPCPEE149 pKa = 4.19MLRR152 pKa = 11.84TLDD155 pKa = 3.31VSGRR159 pKa = 11.84TTLLYY164 pKa = 9.85PSRR167 pKa = 11.84YY168 pKa = 8.46LHH170 pKa = 6.78ARR172 pKa = 11.84DD173 pKa = 3.26RR174 pKa = 11.84MQVEE178 pKa = 4.38KK179 pKa = 10.51TMDD182 pKa = 3.25TAKK185 pKa = 10.6CVSTSVNLDD194 pKa = 3.17ATVVFSVDD202 pKa = 3.76FPFSEE207 pKa = 4.29RR208 pKa = 11.84AILRR212 pKa = 11.84VTQTFYY218 pKa = 10.99PDD220 pKa = 4.09DD221 pKa = 4.31RR222 pKa = 11.84PQKK225 pKa = 10.03HH226 pKa = 6.33LLYY229 pKa = 10.46LASQGGKK236 pKa = 6.3TVHH239 pKa = 5.93YY240 pKa = 9.7SEE242 pKa = 5.31VSATVTPLPGGNASFNVTIEE262 pKa = 4.26FTDD265 pKa = 3.63QPDD268 pKa = 3.48DD269 pKa = 3.76TTIHH273 pKa = 6.25LWVSPKK279 pKa = 10.6GINDD283 pKa = 3.81DD284 pKa = 3.77PDD286 pKa = 3.73AVSAYY291 pKa = 10.5YY292 pKa = 9.98ILNSLSSYY300 pKa = 10.27DD301 pKa = 3.7DD302 pKa = 4.34PYY304 pKa = 11.04AHH306 pKa = 7.22EE307 pKa = 5.43EE308 pKa = 3.97ISCDD312 pKa = 3.18SVYY315 pKa = 11.19QNIVMVLGSVVNITSPDD332 pKa = 3.88PLPRR336 pKa = 11.84APDD339 pKa = 3.38YY340 pKa = 11.14SYY342 pKa = 8.83PTDD345 pKa = 4.01DD346 pKa = 4.17PAGEE350 pKa = 4.33TTEE353 pKa = 5.03SAVTWLEE360 pKa = 3.83ATAQTADD367 pKa = 3.53EE368 pKa = 4.52ATTPLAAHH376 pKa = 6.28ATDD379 pKa = 4.67GYY381 pKa = 11.16EE382 pKa = 4.05PTTAAATLSTTEE394 pKa = 5.34DD395 pKa = 3.35LTQEE399 pKa = 4.1TSTPTVTTVIDD410 pKa = 3.85PTSGAVTTEE419 pKa = 3.52SRR421 pKa = 11.84TTEE424 pKa = 3.65GTAANVATTEE434 pKa = 3.94AAGTEE439 pKa = 4.34GQNQEE444 pKa = 3.77ATTAGGPTNAATTLGHH460 pKa = 5.1QTIEE464 pKa = 3.96AATVEE469 pKa = 4.52DD470 pKa = 3.99STTTARR476 pKa = 11.84AAEE479 pKa = 4.57YY480 pKa = 7.18PTPTTTTVEE489 pKa = 3.96PRR491 pKa = 11.84PAGGTTIEE499 pKa = 5.25DD500 pKa = 4.01PTDD503 pKa = 3.42SPAIEE508 pKa = 4.73DD509 pKa = 3.88TTTPTTTAAKK519 pKa = 8.82VTALKK524 pKa = 9.03TAAAEE529 pKa = 4.57GPTPCVTTYY538 pKa = 10.04TGSDD542 pKa = 3.19TEE544 pKa = 4.4AAQSATSISDD554 pKa = 3.11AVTPEE559 pKa = 4.15DD560 pKa = 4.11LTPEE564 pKa = 4.05TTTPVDD570 pKa = 3.12WSVTSTYY577 pKa = 10.7ISVADD582 pKa = 4.24GTSMPAPTPSAVAHH596 pKa = 6.01EE597 pKa = 4.99ASTTPAPTATAVPTSHH613 pKa = 6.41TPKK616 pKa = 9.38PQEE619 pKa = 4.15STSTPSRR626 pKa = 11.84APTTGVAPVTANSLSPAATSSEE648 pKa = 4.09RR649 pKa = 11.84IVILNTATASSGPGASTGATTAPISPPWSASPAGDD684 pKa = 3.34GVTTSAARR692 pKa = 11.84TLEE695 pKa = 4.05PSSTRR700 pKa = 11.84KK701 pKa = 9.66AVAAEE706 pKa = 4.13STTAADD712 pKa = 4.41DD713 pKa = 3.8VATSEE718 pKa = 4.92LGSGDD723 pKa = 4.02YY724 pKa = 10.71GDD726 pKa = 5.1HH727 pKa = 5.77ATEE730 pKa = 4.32PPRR733 pKa = 11.84IVITNPPGITTLHH746 pKa = 6.78DD747 pKa = 3.78ADD749 pKa = 4.81AAEE752 pKa = 4.43EE753 pKa = 4.63DD754 pKa = 3.81PWPTRR759 pKa = 11.84PLYY762 pKa = 10.67SVNIVNATLTANGMLTATCMAAAKK786 pKa = 9.38AKK788 pKa = 10.28HH789 pKa = 6.34AITFTWHH796 pKa = 5.63VGSNMVPAITGPPEE810 pKa = 4.42PGLMFNGNRR819 pKa = 11.84AWSSRR824 pKa = 11.84LQTVEE829 pKa = 3.96YY830 pKa = 9.92GISPSARR837 pKa = 11.84LACMACTVPPAQRR850 pKa = 11.84YY851 pKa = 6.88CAHH854 pKa = 7.07DD855 pKa = 3.55VAVVARR861 pKa = 11.84HH862 pKa = 6.44DD863 pKa = 3.85RR864 pKa = 11.84LEE866 pKa = 5.66LDD868 pKa = 3.51MQVDD872 pKa = 4.12VATVSVVCSGLDD884 pKa = 3.53GVEE887 pKa = 4.05SEE889 pKa = 5.42PYY891 pKa = 10.11FVWTANGRR899 pKa = 11.84PVPLGSVRR907 pKa = 11.84TKK909 pKa = 10.65RR910 pKa = 11.84IPNDD914 pKa = 3.17YY915 pKa = 7.85GTPARR920 pKa = 11.84WQSAIHH926 pKa = 6.45ISRR929 pKa = 11.84FFVPAGHH936 pKa = 6.76RR937 pKa = 11.84DD938 pKa = 3.6VYY940 pKa = 11.22EE941 pKa = 4.26CTATLASGEE950 pKa = 4.38TIKK953 pKa = 10.26ATKK956 pKa = 9.46NWSNTDD962 pKa = 3.37YY963 pKa = 11.46LALQKK968 pKa = 10.62NAAARR973 pKa = 11.84STFVVAGGITAFAVAEE989 pKa = 4.12LLL991 pKa = 4.14
MM1 pKa = 7.71LWPAALVAMFALAARR16 pKa = 11.84ARR18 pKa = 11.84EE19 pKa = 4.04IDD21 pKa = 3.64NVTCSVVYY29 pKa = 9.66GSNVARR35 pKa = 11.84ISKK38 pKa = 9.95DD39 pKa = 2.97YY40 pKa = 10.5WLAQEE45 pKa = 4.73GSLVSFMTFEE55 pKa = 4.43NDD57 pKa = 3.13DD58 pKa = 3.47PVYY61 pKa = 10.82FFMGRR66 pKa = 11.84ALGTGASGGKK76 pKa = 9.21YY77 pKa = 9.89LYY79 pKa = 10.49RR80 pKa = 11.84VTRR83 pKa = 11.84DD84 pKa = 2.89NNTNVSHH91 pKa = 5.86VQTSLFIPEE100 pKa = 4.62HH101 pKa = 5.8FNGGILLDD109 pKa = 3.48MGSNYY114 pKa = 8.76THH116 pKa = 7.6DD117 pKa = 4.73AEE119 pKa = 4.61TDD121 pKa = 3.68PEE123 pKa = 4.27FLNARR128 pKa = 11.84YY129 pKa = 9.16LVVNDD134 pKa = 4.1TEE136 pKa = 4.73DD137 pKa = 3.35SVRR140 pKa = 11.84EE141 pKa = 4.05EE142 pKa = 4.17TLGPCPEE149 pKa = 4.19MLRR152 pKa = 11.84TLDD155 pKa = 3.31VSGRR159 pKa = 11.84TTLLYY164 pKa = 9.85PSRR167 pKa = 11.84YY168 pKa = 8.46LHH170 pKa = 6.78ARR172 pKa = 11.84DD173 pKa = 3.26RR174 pKa = 11.84MQVEE178 pKa = 4.38KK179 pKa = 10.51TMDD182 pKa = 3.25TAKK185 pKa = 10.6CVSTSVNLDD194 pKa = 3.17ATVVFSVDD202 pKa = 3.76FPFSEE207 pKa = 4.29RR208 pKa = 11.84AILRR212 pKa = 11.84VTQTFYY218 pKa = 10.99PDD220 pKa = 4.09DD221 pKa = 4.31RR222 pKa = 11.84PQKK225 pKa = 10.03HH226 pKa = 6.33LLYY229 pKa = 10.46LASQGGKK236 pKa = 6.3TVHH239 pKa = 5.93YY240 pKa = 9.7SEE242 pKa = 5.31VSATVTPLPGGNASFNVTIEE262 pKa = 4.26FTDD265 pKa = 3.63QPDD268 pKa = 3.48DD269 pKa = 3.76TTIHH273 pKa = 6.25LWVSPKK279 pKa = 10.6GINDD283 pKa = 3.81DD284 pKa = 3.77PDD286 pKa = 3.73AVSAYY291 pKa = 10.5YY292 pKa = 9.98ILNSLSSYY300 pKa = 10.27DD301 pKa = 3.7DD302 pKa = 4.34PYY304 pKa = 11.04AHH306 pKa = 7.22EE307 pKa = 5.43EE308 pKa = 3.97ISCDD312 pKa = 3.18SVYY315 pKa = 11.19QNIVMVLGSVVNITSPDD332 pKa = 3.88PLPRR336 pKa = 11.84APDD339 pKa = 3.38YY340 pKa = 11.14SYY342 pKa = 8.83PTDD345 pKa = 4.01DD346 pKa = 4.17PAGEE350 pKa = 4.33TTEE353 pKa = 5.03SAVTWLEE360 pKa = 3.83ATAQTADD367 pKa = 3.53EE368 pKa = 4.52ATTPLAAHH376 pKa = 6.28ATDD379 pKa = 4.67GYY381 pKa = 11.16EE382 pKa = 4.05PTTAAATLSTTEE394 pKa = 5.34DD395 pKa = 3.35LTQEE399 pKa = 4.1TSTPTVTTVIDD410 pKa = 3.85PTSGAVTTEE419 pKa = 3.52SRR421 pKa = 11.84TTEE424 pKa = 3.65GTAANVATTEE434 pKa = 3.94AAGTEE439 pKa = 4.34GQNQEE444 pKa = 3.77ATTAGGPTNAATTLGHH460 pKa = 5.1QTIEE464 pKa = 3.96AATVEE469 pKa = 4.52DD470 pKa = 3.99STTTARR476 pKa = 11.84AAEE479 pKa = 4.57YY480 pKa = 7.18PTPTTTTVEE489 pKa = 3.96PRR491 pKa = 11.84PAGGTTIEE499 pKa = 5.25DD500 pKa = 4.01PTDD503 pKa = 3.42SPAIEE508 pKa = 4.73DD509 pKa = 3.88TTTPTTTAAKK519 pKa = 8.82VTALKK524 pKa = 9.03TAAAEE529 pKa = 4.57GPTPCVTTYY538 pKa = 10.04TGSDD542 pKa = 3.19TEE544 pKa = 4.4AAQSATSISDD554 pKa = 3.11AVTPEE559 pKa = 4.15DD560 pKa = 4.11LTPEE564 pKa = 4.05TTTPVDD570 pKa = 3.12WSVTSTYY577 pKa = 10.7ISVADD582 pKa = 4.24GTSMPAPTPSAVAHH596 pKa = 6.01EE597 pKa = 4.99ASTTPAPTATAVPTSHH613 pKa = 6.41TPKK616 pKa = 9.38PQEE619 pKa = 4.15STSTPSRR626 pKa = 11.84APTTGVAPVTANSLSPAATSSEE648 pKa = 4.09RR649 pKa = 11.84IVILNTATASSGPGASTGATTAPISPPWSASPAGDD684 pKa = 3.34GVTTSAARR692 pKa = 11.84TLEE695 pKa = 4.05PSSTRR700 pKa = 11.84KK701 pKa = 9.66AVAAEE706 pKa = 4.13STTAADD712 pKa = 4.41DD713 pKa = 3.8VATSEE718 pKa = 4.92LGSGDD723 pKa = 4.02YY724 pKa = 10.71GDD726 pKa = 5.1HH727 pKa = 5.77ATEE730 pKa = 4.32PPRR733 pKa = 11.84IVITNPPGITTLHH746 pKa = 6.78DD747 pKa = 3.78ADD749 pKa = 4.81AAEE752 pKa = 4.43EE753 pKa = 4.63DD754 pKa = 3.81PWPTRR759 pKa = 11.84PLYY762 pKa = 10.67SVNIVNATLTANGMLTATCMAAAKK786 pKa = 9.38AKK788 pKa = 10.28HH789 pKa = 6.34AITFTWHH796 pKa = 5.63VGSNMVPAITGPPEE810 pKa = 4.42PGLMFNGNRR819 pKa = 11.84AWSSRR824 pKa = 11.84LQTVEE829 pKa = 3.96YY830 pKa = 9.92GISPSARR837 pKa = 11.84LACMACTVPPAQRR850 pKa = 11.84YY851 pKa = 6.88CAHH854 pKa = 7.07DD855 pKa = 3.55VAVVARR861 pKa = 11.84HH862 pKa = 6.44DD863 pKa = 3.85RR864 pKa = 11.84LEE866 pKa = 5.66LDD868 pKa = 3.51MQVDD872 pKa = 4.12VATVSVVCSGLDD884 pKa = 3.53GVEE887 pKa = 4.05SEE889 pKa = 5.42PYY891 pKa = 10.11FVWTANGRR899 pKa = 11.84PVPLGSVRR907 pKa = 11.84TKK909 pKa = 10.65RR910 pKa = 11.84IPNDD914 pKa = 3.17YY915 pKa = 7.85GTPARR920 pKa = 11.84WQSAIHH926 pKa = 6.45ISRR929 pKa = 11.84FFVPAGHH936 pKa = 6.76RR937 pKa = 11.84DD938 pKa = 3.6VYY940 pKa = 11.22EE941 pKa = 4.26CTATLASGEE950 pKa = 4.38TIKK953 pKa = 10.26ATKK956 pKa = 9.46NWSNTDD962 pKa = 3.37YY963 pKa = 11.46LALQKK968 pKa = 10.62NAAARR973 pKa = 11.84STFVVAGGITAFAVAEE989 pKa = 4.12LLL991 pKa = 4.14
Molecular weight: 104.42 kDa
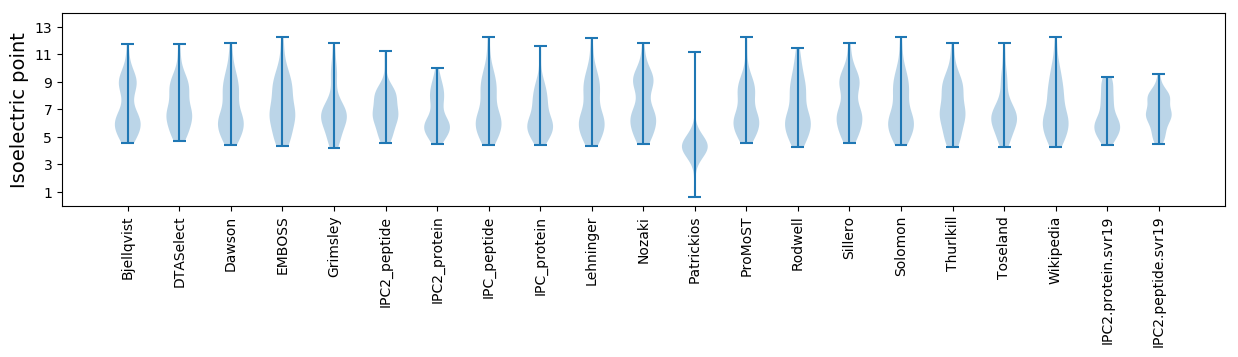
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q6UDK9|KITH_PSHV1 Thymidine kinase OS=Psittacid herpesvirus 1 (isolate Amazon parrot/-/97-0001/1997) OX=670426 GN=TK PE=3 SV=1
MM1 pKa = 7.39PRR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84GPRR9 pKa = 11.84CDD11 pKa = 3.54PSSALAVDD19 pKa = 3.95VAAAVAAATSRR30 pKa = 11.84TRR32 pKa = 11.84AQKK35 pKa = 9.49VRR37 pKa = 11.84KK38 pKa = 8.76AKK40 pKa = 10.25KK41 pKa = 8.15VLAANGHH48 pKa = 5.21KK49 pKa = 8.31WHH51 pKa = 6.99RR52 pKa = 11.84RR53 pKa = 11.84AKK55 pKa = 8.93TPVNKK60 pKa = 8.94RR61 pKa = 11.84TLVAARR67 pKa = 11.84LLAGVRR73 pKa = 11.84CHH75 pKa = 5.73NRR77 pKa = 11.84FYY79 pKa = 11.26GALVSSFEE87 pKa = 3.96TAYY90 pKa = 11.04CMNHH94 pKa = 4.89RR95 pKa = 11.84HH96 pKa = 5.85GSDD99 pKa = 2.98GLAATIIAEE108 pKa = 4.19ACGAVPVCSLNTTIMFEE125 pKa = 4.22VNLGGRR131 pKa = 11.84RR132 pKa = 11.84PDD134 pKa = 4.31CICVLDD140 pKa = 3.85RR141 pKa = 11.84AGVAGAPRR149 pKa = 11.84EE150 pKa = 4.36SVCLIVEE157 pKa = 4.72LKK159 pKa = 8.72TCRR162 pKa = 11.84FSRR165 pKa = 11.84SALTEE170 pKa = 4.14SKK172 pKa = 10.4KK173 pKa = 10.38NQYY176 pKa = 8.5ATGLRR181 pKa = 11.84QLRR184 pKa = 11.84DD185 pKa = 3.36SAKK188 pKa = 9.91IICDD192 pKa = 3.45VAVPGADD199 pKa = 3.05FVHH202 pKa = 6.57IVPVLVFVAQRR213 pKa = 11.84SMRR216 pKa = 11.84IMDD219 pKa = 3.03VRR221 pKa = 11.84KK222 pKa = 8.99FNEE225 pKa = 3.8RR226 pKa = 11.84VVRR229 pKa = 11.84ANAEE233 pKa = 4.18LLRR236 pKa = 11.84LRR238 pKa = 11.84LAALGVYY245 pKa = 9.63SRR247 pKa = 11.84SFAAAPQRR255 pKa = 11.84LQSAQDD261 pKa = 3.2RR262 pKa = 11.84CHH264 pKa = 6.78DD265 pKa = 3.68HH266 pKa = 7.13AATPKK271 pKa = 8.0PTPAVVPSPPAPPNPAPPQSRR292 pKa = 11.84VGFGSALAMATAVMLGEE309 pKa = 4.21YY310 pKa = 10.27ARR312 pKa = 11.84EE313 pKa = 4.06NKK315 pKa = 9.73PPRR318 pKa = 4.13
MM1 pKa = 7.39PRR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84GPRR9 pKa = 11.84CDD11 pKa = 3.54PSSALAVDD19 pKa = 3.95VAAAVAAATSRR30 pKa = 11.84TRR32 pKa = 11.84AQKK35 pKa = 9.49VRR37 pKa = 11.84KK38 pKa = 8.76AKK40 pKa = 10.25KK41 pKa = 8.15VLAANGHH48 pKa = 5.21KK49 pKa = 8.31WHH51 pKa = 6.99RR52 pKa = 11.84RR53 pKa = 11.84AKK55 pKa = 8.93TPVNKK60 pKa = 8.94RR61 pKa = 11.84TLVAARR67 pKa = 11.84LLAGVRR73 pKa = 11.84CHH75 pKa = 5.73NRR77 pKa = 11.84FYY79 pKa = 11.26GALVSSFEE87 pKa = 3.96TAYY90 pKa = 11.04CMNHH94 pKa = 4.89RR95 pKa = 11.84HH96 pKa = 5.85GSDD99 pKa = 2.98GLAATIIAEE108 pKa = 4.19ACGAVPVCSLNTTIMFEE125 pKa = 4.22VNLGGRR131 pKa = 11.84RR132 pKa = 11.84PDD134 pKa = 4.31CICVLDD140 pKa = 3.85RR141 pKa = 11.84AGVAGAPRR149 pKa = 11.84EE150 pKa = 4.36SVCLIVEE157 pKa = 4.72LKK159 pKa = 8.72TCRR162 pKa = 11.84FSRR165 pKa = 11.84SALTEE170 pKa = 4.14SKK172 pKa = 10.4KK173 pKa = 10.38NQYY176 pKa = 8.5ATGLRR181 pKa = 11.84QLRR184 pKa = 11.84DD185 pKa = 3.36SAKK188 pKa = 9.91IICDD192 pKa = 3.45VAVPGADD199 pKa = 3.05FVHH202 pKa = 6.57IVPVLVFVAQRR213 pKa = 11.84SMRR216 pKa = 11.84IMDD219 pKa = 3.03VRR221 pKa = 11.84KK222 pKa = 8.99FNEE225 pKa = 3.8RR226 pKa = 11.84VVRR229 pKa = 11.84ANAEE233 pKa = 4.18LLRR236 pKa = 11.84LRR238 pKa = 11.84LAALGVYY245 pKa = 9.63SRR247 pKa = 11.84SFAAAPQRR255 pKa = 11.84LQSAQDD261 pKa = 3.2RR262 pKa = 11.84CHH264 pKa = 6.78DD265 pKa = 3.68HH266 pKa = 7.13AATPKK271 pKa = 8.0PTPAVVPSPPAPPNPAPPQSRR292 pKa = 11.84VGFGSALAMATAVMLGEE309 pKa = 4.21YY310 pKa = 10.27ARR312 pKa = 11.84EE313 pKa = 4.06NKK315 pKa = 9.73PPRR318 pKa = 4.13
Molecular weight: 34.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
40367 |
47 |
3195 |
560.7 |
61.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.906 ± 0.355 | 2.049 ± 0.126 |
5.757 ± 0.23 | 5.854 ± 0.238 |
3.651 ± 0.169 | 6.934 ± 0.178 |
2.059 ± 0.074 | 3.647 ± 0.137 |
3.518 ± 0.209 | 9.084 ± 0.221 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.356 ± 0.121 | 2.918 ± 0.129 |
5.965 ± 0.297 | 2.75 ± 0.13 |
7.62 ± 0.252 | 7.578 ± 0.235 |
5.638 ± 0.291 | 6.76 ± 0.208 |
1.147 ± 0.051 | 2.809 ± 0.132 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
