
Mesorhizobium oceanicum
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Phyllobacteriaceae; Mesorhizobium
Average proteome isoelectric point is 6.31
Get precalculated fractions of proteins
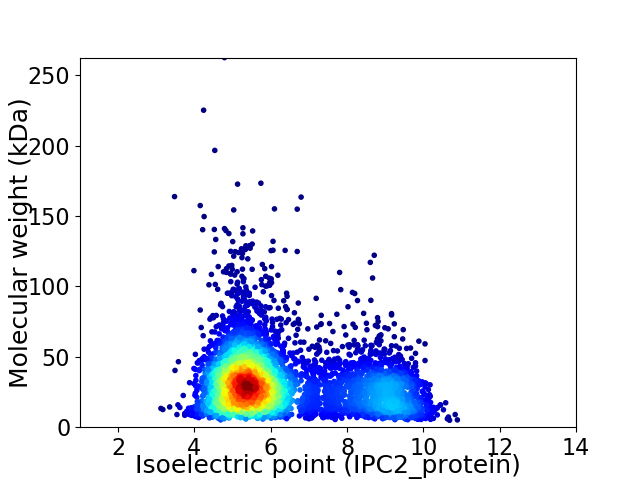
Virtual 2D-PAGE plot for 5014 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3SN42|A0A1L3SN42_9RHIZ ABC transporter substrate-binding protein OS=Mesorhizobium oceanicum OX=1670800 GN=BSQ44_04520 PE=4 SV=1
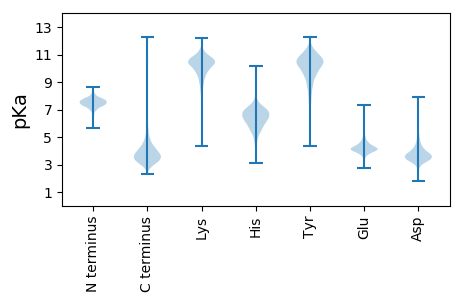
MM1 pKa = 7.95RR2 pKa = 11.84IANTSTALALLMTLGAGPAFAAAEE26 pKa = 4.22WTFSDD31 pKa = 4.38IDD33 pKa = 3.4QDD35 pKa = 4.53GNLEE39 pKa = 4.02LSNKK43 pKa = 8.72EE44 pKa = 3.96FEE46 pKa = 4.46QVSQGAFRR54 pKa = 11.84SWDD57 pKa = 3.49TNQDD61 pKa = 2.93QRR63 pKa = 11.84LTSDD67 pKa = 3.13EE68 pKa = 4.39LDD70 pKa = 3.46RR71 pKa = 11.84GVYY74 pKa = 10.42ASWDD78 pKa = 3.44RR79 pKa = 11.84DD80 pKa = 3.22RR81 pKa = 11.84DD82 pKa = 3.78DD83 pKa = 4.67RR84 pKa = 11.84LSEE87 pKa = 4.34SEE89 pKa = 5.42FDD91 pKa = 4.02SGWLGWFGDD100 pKa = 4.41DD101 pKa = 5.03DD102 pKa = 3.84QVAYY106 pKa = 11.02SDD108 pKa = 4.31LDD110 pKa = 4.16RR111 pKa = 11.84NGDD114 pKa = 3.39AYY116 pKa = 11.24LDD118 pKa = 3.87EE119 pKa = 6.47DD120 pKa = 4.44EE121 pKa = 4.93FTAGLQQSAALDD133 pKa = 3.57DD134 pKa = 4.23WNVGDD139 pKa = 5.97EE140 pKa = 4.36GVDD143 pKa = 3.06WQNFHH148 pKa = 5.97TALYY152 pKa = 10.22GVYY155 pKa = 9.15DD156 pKa = 3.72TDD158 pKa = 3.67KK159 pKa = 11.15NAMVNQDD166 pKa = 3.11EE167 pKa = 4.49YY168 pKa = 11.71GAYY171 pKa = 10.23SDD173 pKa = 3.95TYY175 pKa = 10.97MMGDD179 pKa = 3.47MAQAGVGAEE188 pKa = 3.82EE189 pKa = 3.97TAAVGEE195 pKa = 4.6TITPQEE201 pKa = 4.6VISMSDD207 pKa = 2.82WRR209 pKa = 11.84SEE211 pKa = 3.73DD212 pKa = 3.68LYY214 pKa = 11.24LGGMSVDD221 pKa = 3.35EE222 pKa = 4.85MMDD225 pKa = 3.32DD226 pKa = 3.7MEE228 pKa = 5.09VYY230 pKa = 10.56GPTGEE235 pKa = 4.66EE236 pKa = 3.56IGSVEE241 pKa = 3.93NVVFSNDD248 pKa = 2.74GRR250 pKa = 11.84VLSLVAEE257 pKa = 4.43VGGFWDD263 pKa = 4.07MFDD266 pKa = 3.33THH268 pKa = 7.68VNVPWDD274 pKa = 3.4QVNYY278 pKa = 10.36SGEE281 pKa = 4.11DD282 pKa = 3.37KK283 pKa = 10.74IIIPVTEE290 pKa = 4.44EE291 pKa = 3.69NVEE294 pKa = 5.14DD295 pKa = 3.93YY296 pKa = 9.94STWKK300 pKa = 9.15TGYY303 pKa = 7.89LTPQDD308 pKa = 3.45AMGVEE313 pKa = 4.61VVDD316 pKa = 5.58DD317 pKa = 4.6DD318 pKa = 6.05LEE320 pKa = 4.34TSPGIFRR327 pKa = 11.84ATDD330 pKa = 3.17LMGDD334 pKa = 3.62YY335 pKa = 11.07ARR337 pKa = 11.84VKK339 pKa = 10.21GGEE342 pKa = 3.9ANGYY346 pKa = 9.54SNYY349 pKa = 10.51GYY351 pKa = 11.31VNDD354 pKa = 5.42LIIRR358 pKa = 11.84DD359 pKa = 3.96GQLQAVVVSPDD370 pKa = 2.91AGYY373 pKa = 10.73GISGPYY379 pKa = 9.52AYY381 pKa = 9.36PYY383 pKa = 10.49YY384 pKa = 10.56SRR386 pKa = 11.84GWSAGRR392 pKa = 11.84PYY394 pKa = 11.39YY395 pKa = 10.62DD396 pKa = 3.36MPYY399 pKa = 10.94DD400 pKa = 3.73RR401 pKa = 11.84ADD403 pKa = 3.49VVDD406 pKa = 5.41NEE408 pKa = 4.16PLDD411 pKa = 3.72YY412 pKa = 11.3DD413 pKa = 3.68RR414 pKa = 11.84FGVQQ418 pKa = 3.2
MM1 pKa = 7.95RR2 pKa = 11.84IANTSTALALLMTLGAGPAFAAAEE26 pKa = 4.22WTFSDD31 pKa = 4.38IDD33 pKa = 3.4QDD35 pKa = 4.53GNLEE39 pKa = 4.02LSNKK43 pKa = 8.72EE44 pKa = 3.96FEE46 pKa = 4.46QVSQGAFRR54 pKa = 11.84SWDD57 pKa = 3.49TNQDD61 pKa = 2.93QRR63 pKa = 11.84LTSDD67 pKa = 3.13EE68 pKa = 4.39LDD70 pKa = 3.46RR71 pKa = 11.84GVYY74 pKa = 10.42ASWDD78 pKa = 3.44RR79 pKa = 11.84DD80 pKa = 3.22RR81 pKa = 11.84DD82 pKa = 3.78DD83 pKa = 4.67RR84 pKa = 11.84LSEE87 pKa = 4.34SEE89 pKa = 5.42FDD91 pKa = 4.02SGWLGWFGDD100 pKa = 4.41DD101 pKa = 5.03DD102 pKa = 3.84QVAYY106 pKa = 11.02SDD108 pKa = 4.31LDD110 pKa = 4.16RR111 pKa = 11.84NGDD114 pKa = 3.39AYY116 pKa = 11.24LDD118 pKa = 3.87EE119 pKa = 6.47DD120 pKa = 4.44EE121 pKa = 4.93FTAGLQQSAALDD133 pKa = 3.57DD134 pKa = 4.23WNVGDD139 pKa = 5.97EE140 pKa = 4.36GVDD143 pKa = 3.06WQNFHH148 pKa = 5.97TALYY152 pKa = 10.22GVYY155 pKa = 9.15DD156 pKa = 3.72TDD158 pKa = 3.67KK159 pKa = 11.15NAMVNQDD166 pKa = 3.11EE167 pKa = 4.49YY168 pKa = 11.71GAYY171 pKa = 10.23SDD173 pKa = 3.95TYY175 pKa = 10.97MMGDD179 pKa = 3.47MAQAGVGAEE188 pKa = 3.82EE189 pKa = 3.97TAAVGEE195 pKa = 4.6TITPQEE201 pKa = 4.6VISMSDD207 pKa = 2.82WRR209 pKa = 11.84SEE211 pKa = 3.73DD212 pKa = 3.68LYY214 pKa = 11.24LGGMSVDD221 pKa = 3.35EE222 pKa = 4.85MMDD225 pKa = 3.32DD226 pKa = 3.7MEE228 pKa = 5.09VYY230 pKa = 10.56GPTGEE235 pKa = 4.66EE236 pKa = 3.56IGSVEE241 pKa = 3.93NVVFSNDD248 pKa = 2.74GRR250 pKa = 11.84VLSLVAEE257 pKa = 4.43VGGFWDD263 pKa = 4.07MFDD266 pKa = 3.33THH268 pKa = 7.68VNVPWDD274 pKa = 3.4QVNYY278 pKa = 10.36SGEE281 pKa = 4.11DD282 pKa = 3.37KK283 pKa = 10.74IIIPVTEE290 pKa = 4.44EE291 pKa = 3.69NVEE294 pKa = 5.14DD295 pKa = 3.93YY296 pKa = 9.94STWKK300 pKa = 9.15TGYY303 pKa = 7.89LTPQDD308 pKa = 3.45AMGVEE313 pKa = 4.61VVDD316 pKa = 5.58DD317 pKa = 4.6DD318 pKa = 6.05LEE320 pKa = 4.34TSPGIFRR327 pKa = 11.84ATDD330 pKa = 3.17LMGDD334 pKa = 3.62YY335 pKa = 11.07ARR337 pKa = 11.84VKK339 pKa = 10.21GGEE342 pKa = 3.9ANGYY346 pKa = 9.54SNYY349 pKa = 10.51GYY351 pKa = 11.31VNDD354 pKa = 5.42LIIRR358 pKa = 11.84DD359 pKa = 3.96GQLQAVVVSPDD370 pKa = 2.91AGYY373 pKa = 10.73GISGPYY379 pKa = 9.52AYY381 pKa = 9.36PYY383 pKa = 10.49YY384 pKa = 10.56SRR386 pKa = 11.84GWSAGRR392 pKa = 11.84PYY394 pKa = 11.39YY395 pKa = 10.62DD396 pKa = 3.36MPYY399 pKa = 10.94DD400 pKa = 3.73RR401 pKa = 11.84ADD403 pKa = 3.49VVDD406 pKa = 5.41NEE408 pKa = 4.16PLDD411 pKa = 3.72YY412 pKa = 11.3DD413 pKa = 3.68RR414 pKa = 11.84FGVQQ418 pKa = 3.2
Molecular weight: 46.51 kDa
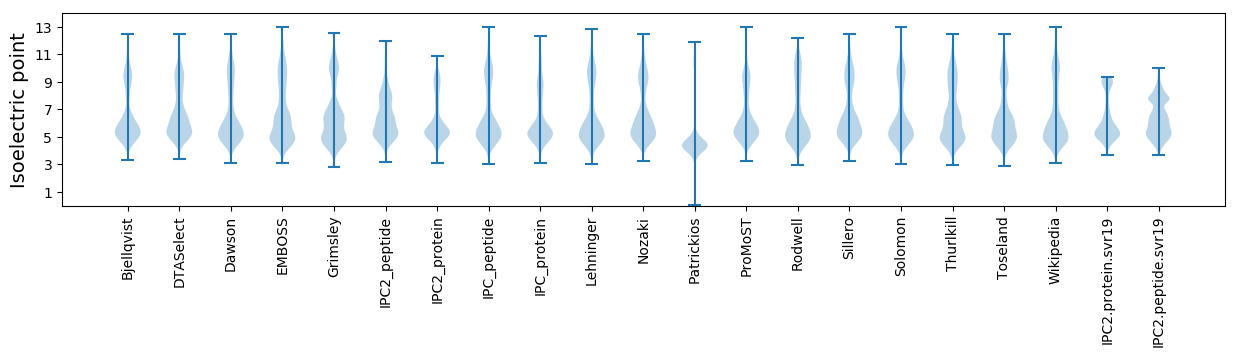
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3SVM4|A0A1L3SVM4_9RHIZ Voltage-gated sodium channel OS=Mesorhizobium oceanicum OX=1670800 GN=BSQ44_20185 PE=4 SV=1
MM1 pKa = 7.32NRR3 pKa = 11.84TTPLVLAVALFMEE16 pKa = 4.76QMDD19 pKa = 3.98STVIATSLPAIAADD33 pKa = 3.65IEE35 pKa = 4.62TNPIALKK42 pKa = 10.55LALTAYY48 pKa = 9.23LVSVAIFIPISAWMADD64 pKa = 3.17RR65 pKa = 11.84FGAKK69 pKa = 8.91RR70 pKa = 11.84VFCTAIGVFVLGSIACAMASSLGEE94 pKa = 3.73FVLARR99 pKa = 11.84FLQGMGGSMMTPVARR114 pKa = 11.84LLLVRR119 pKa = 11.84ATPRR123 pKa = 11.84SNLVSAMAWLTVPALVGPLVGPPVGGFITTYY154 pKa = 10.8VSWHH158 pKa = 6.57WIFLINVPIGITGICAAWLLLPEE181 pKa = 4.83LEE183 pKa = 4.3RR184 pKa = 11.84GIAGRR189 pKa = 11.84IDD191 pKa = 3.5FTGFALSGMAAAGIVFGLSVVSLPALPPAVGVATLALGICSGLLYY236 pKa = 10.17FRR238 pKa = 11.84HH239 pKa = 6.07ARR241 pKa = 11.84RR242 pKa = 11.84VARR245 pKa = 11.84PILDD249 pKa = 3.34LSLFRR254 pKa = 11.84NRR256 pKa = 11.84VFRR259 pKa = 11.84AAISGGSLFRR269 pKa = 11.84IGAGATPFLLPLMFQLGFGLTPFQSGMLTFAGAFGAIGMKK309 pKa = 10.15FFASTTLRR317 pKa = 11.84KK318 pKa = 10.16GGFRR322 pKa = 11.84TVLISTAIAGGIFIGVAAAFTAQTPAWAIIAVLVAGGFLRR362 pKa = 11.84SLFFTSANALVFADD376 pKa = 4.35IGDD379 pKa = 3.88EE380 pKa = 4.08RR381 pKa = 11.84AGQATAIAAVTQQISIALGVALAGGILEE409 pKa = 4.31GVAMFSGDD417 pKa = 3.5TLGASAFPIAFVLVGLVTTLAALPFLTLPPDD448 pKa = 3.77AGSAVSGHH456 pKa = 6.21RR457 pKa = 11.84VDD459 pKa = 4.37GVNSRR464 pKa = 11.84TQVVPAKK471 pKa = 10.57
MM1 pKa = 7.32NRR3 pKa = 11.84TTPLVLAVALFMEE16 pKa = 4.76QMDD19 pKa = 3.98STVIATSLPAIAADD33 pKa = 3.65IEE35 pKa = 4.62TNPIALKK42 pKa = 10.55LALTAYY48 pKa = 9.23LVSVAIFIPISAWMADD64 pKa = 3.17RR65 pKa = 11.84FGAKK69 pKa = 8.91RR70 pKa = 11.84VFCTAIGVFVLGSIACAMASSLGEE94 pKa = 3.73FVLARR99 pKa = 11.84FLQGMGGSMMTPVARR114 pKa = 11.84LLLVRR119 pKa = 11.84ATPRR123 pKa = 11.84SNLVSAMAWLTVPALVGPLVGPPVGGFITTYY154 pKa = 10.8VSWHH158 pKa = 6.57WIFLINVPIGITGICAAWLLLPEE181 pKa = 4.83LEE183 pKa = 4.3RR184 pKa = 11.84GIAGRR189 pKa = 11.84IDD191 pKa = 3.5FTGFALSGMAAAGIVFGLSVVSLPALPPAVGVATLALGICSGLLYY236 pKa = 10.17FRR238 pKa = 11.84HH239 pKa = 6.07ARR241 pKa = 11.84RR242 pKa = 11.84VARR245 pKa = 11.84PILDD249 pKa = 3.34LSLFRR254 pKa = 11.84NRR256 pKa = 11.84VFRR259 pKa = 11.84AAISGGSLFRR269 pKa = 11.84IGAGATPFLLPLMFQLGFGLTPFQSGMLTFAGAFGAIGMKK309 pKa = 10.15FFASTTLRR317 pKa = 11.84KK318 pKa = 10.16GGFRR322 pKa = 11.84TVLISTAIAGGIFIGVAAAFTAQTPAWAIIAVLVAGGFLRR362 pKa = 11.84SLFFTSANALVFADD376 pKa = 4.35IGDD379 pKa = 3.88EE380 pKa = 4.08RR381 pKa = 11.84AGQATAIAAVTQQISIALGVALAGGILEE409 pKa = 4.31GVAMFSGDD417 pKa = 3.5TLGASAFPIAFVLVGLVTTLAALPFLTLPPDD448 pKa = 3.77AGSAVSGHH456 pKa = 6.21RR457 pKa = 11.84VDD459 pKa = 4.37GVNSRR464 pKa = 11.84TQVVPAKK471 pKa = 10.57
Molecular weight: 48.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1537093 |
41 |
2417 |
306.6 |
33.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.288 ± 0.043 | 0.805 ± 0.008 |
5.779 ± 0.027 | 6.263 ± 0.037 |
3.905 ± 0.025 | 8.731 ± 0.031 |
1.977 ± 0.016 | 5.327 ± 0.026 |
3.228 ± 0.028 | 9.676 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.646 ± 0.017 | 2.506 ± 0.017 |
5.063 ± 0.026 | 2.804 ± 0.02 |
7.241 ± 0.034 | 5.497 ± 0.029 |
5.134 ± 0.027 | 7.552 ± 0.028 |
1.335 ± 0.014 | 2.242 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
