
Ceratocystis fimbriata f. sp. platani
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Hypocreomycetidae; Microascales; Ceratocystidaceae; Ceratocystis
Average proteome isoelectric point is 6.31
Get precalculated fractions of proteins
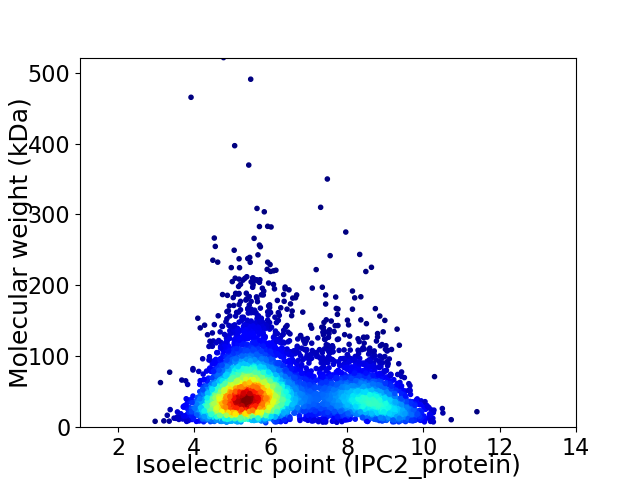
Virtual 2D-PAGE plot for 5620 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
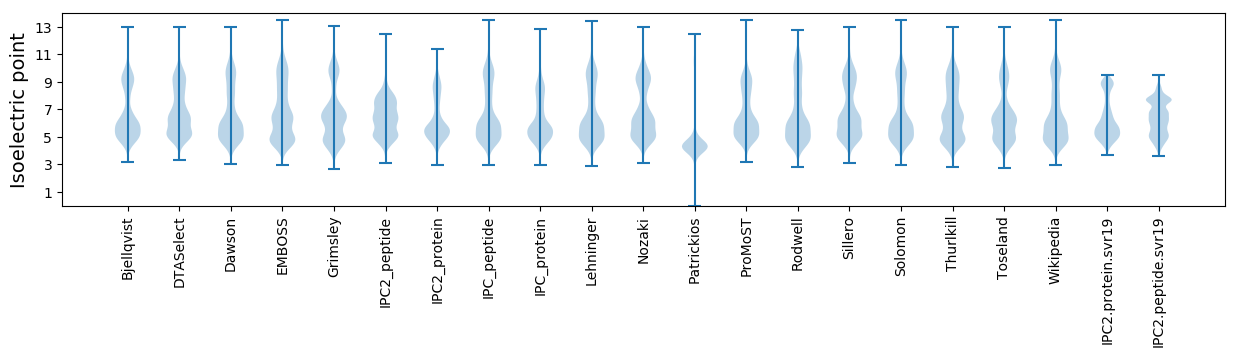
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0F8BTT5|A0A0F8BTT5_CERFI Indoleamine 2 3-dioxygenase OS=Ceratocystis fimbriata f. sp. platani OX=88771 GN=BNA2 PE=3 SV=1
MM1 pKa = 8.14DD2 pKa = 5.65GGTSTDD8 pKa = 3.3GGDD11 pKa = 3.26RR12 pKa = 11.84SNNFTFEE19 pKa = 4.07LMAGDD24 pKa = 5.14DD25 pKa = 3.82EE26 pKa = 6.9SIDD29 pKa = 3.59EE30 pKa = 4.7DD31 pKa = 3.98LNRR34 pKa = 11.84SYY36 pKa = 11.42GRR38 pKa = 11.84QHH40 pKa = 7.0RR41 pKa = 11.84GFEE44 pKa = 4.01NSFDD48 pKa = 3.82SSLSDD53 pKa = 4.65GSDD56 pKa = 3.45PDD58 pKa = 6.01DD59 pKa = 6.72DD60 pKa = 6.99DD61 pKa = 7.77DD62 pKa = 7.79DD63 pKa = 7.51DD64 pKa = 7.04DD65 pKa = 7.3DD66 pKa = 4.88EE67 pKa = 7.04DD68 pKa = 4.98NEE70 pKa = 4.73SSHH73 pKa = 6.84PWPHH77 pKa = 6.15HH78 pKa = 5.81QNDD81 pKa = 4.21FAPPYY86 pKa = 9.18YY87 pKa = 10.39GRR89 pKa = 11.84PPTPLPPSPSITSLLRR105 pKa = 11.84PSRR108 pKa = 11.84PTTPDD113 pKa = 2.95ASDD116 pKa = 4.92DD117 pKa = 3.66EE118 pKa = 4.92LEE120 pKa = 4.42PVPRR124 pKa = 11.84AAPRR128 pKa = 11.84VPTYY132 pKa = 10.26EE133 pKa = 3.77YY134 pKa = 11.02YY135 pKa = 11.26GFVLYY140 pKa = 10.61LFSSLCFLTYY150 pKa = 10.59LLWAFLPSPFLHH162 pKa = 6.81ALGITYY168 pKa = 10.55YY169 pKa = 10.05PDD171 pKa = 3.21RR172 pKa = 11.84WWALAMPTFLVVTLIYY188 pKa = 10.12IYY190 pKa = 10.8VALGSYY196 pKa = 7.87NTEE199 pKa = 3.77ILTLPLTAVEE209 pKa = 4.23TVIDD213 pKa = 4.21DD214 pKa = 4.15AAQVALVDD222 pKa = 3.76AAGSLRR228 pKa = 11.84DD229 pKa = 3.83GSRR232 pKa = 11.84MTPAKK237 pKa = 9.49IDD239 pKa = 3.42VMKK242 pKa = 10.85GYY244 pKa = 11.16GDD246 pKa = 4.65LATIWSEE253 pKa = 3.92GTDD256 pKa = 3.09AVMDD260 pKa = 3.74IPLGAVCEE268 pKa = 4.11VLYY271 pKa = 11.11ARR273 pKa = 11.84GPEE276 pKa = 4.08VFDD279 pKa = 4.34WEE281 pKa = 4.64DD282 pKa = 3.91DD283 pKa = 3.88YY284 pKa = 12.35NEE286 pKa = 3.96
MM1 pKa = 8.14DD2 pKa = 5.65GGTSTDD8 pKa = 3.3GGDD11 pKa = 3.26RR12 pKa = 11.84SNNFTFEE19 pKa = 4.07LMAGDD24 pKa = 5.14DD25 pKa = 3.82EE26 pKa = 6.9SIDD29 pKa = 3.59EE30 pKa = 4.7DD31 pKa = 3.98LNRR34 pKa = 11.84SYY36 pKa = 11.42GRR38 pKa = 11.84QHH40 pKa = 7.0RR41 pKa = 11.84GFEE44 pKa = 4.01NSFDD48 pKa = 3.82SSLSDD53 pKa = 4.65GSDD56 pKa = 3.45PDD58 pKa = 6.01DD59 pKa = 6.72DD60 pKa = 6.99DD61 pKa = 7.77DD62 pKa = 7.79DD63 pKa = 7.51DD64 pKa = 7.04DD65 pKa = 7.3DD66 pKa = 4.88EE67 pKa = 7.04DD68 pKa = 4.98NEE70 pKa = 4.73SSHH73 pKa = 6.84PWPHH77 pKa = 6.15HH78 pKa = 5.81QNDD81 pKa = 4.21FAPPYY86 pKa = 9.18YY87 pKa = 10.39GRR89 pKa = 11.84PPTPLPPSPSITSLLRR105 pKa = 11.84PSRR108 pKa = 11.84PTTPDD113 pKa = 2.95ASDD116 pKa = 4.92DD117 pKa = 3.66EE118 pKa = 4.92LEE120 pKa = 4.42PVPRR124 pKa = 11.84AAPRR128 pKa = 11.84VPTYY132 pKa = 10.26EE133 pKa = 3.77YY134 pKa = 11.02YY135 pKa = 11.26GFVLYY140 pKa = 10.61LFSSLCFLTYY150 pKa = 10.59LLWAFLPSPFLHH162 pKa = 6.81ALGITYY168 pKa = 10.55YY169 pKa = 10.05PDD171 pKa = 3.21RR172 pKa = 11.84WWALAMPTFLVVTLIYY188 pKa = 10.12IYY190 pKa = 10.8VALGSYY196 pKa = 7.87NTEE199 pKa = 3.77ILTLPLTAVEE209 pKa = 4.23TVIDD213 pKa = 4.21DD214 pKa = 4.15AAQVALVDD222 pKa = 3.76AAGSLRR228 pKa = 11.84DD229 pKa = 3.83GSRR232 pKa = 11.84MTPAKK237 pKa = 9.49IDD239 pKa = 3.42VMKK242 pKa = 10.85GYY244 pKa = 11.16GDD246 pKa = 4.65LATIWSEE253 pKa = 3.92GTDD256 pKa = 3.09AVMDD260 pKa = 3.74IPLGAVCEE268 pKa = 4.11VLYY271 pKa = 11.11ARR273 pKa = 11.84GPEE276 pKa = 4.08VFDD279 pKa = 4.34WEE281 pKa = 4.64DD282 pKa = 3.91DD283 pKa = 3.88YY284 pKa = 12.35NEE286 pKa = 3.96
Molecular weight: 31.8 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0F8B468|A0A0F8B468_CERFI Uncharacterized protein OS=Ceratocystis fimbriata f. sp. platani OX=88771 GN=CFO_g1104 PE=4 SV=1
MM1 pKa = 6.86QRR3 pKa = 11.84PRR5 pKa = 11.84RR6 pKa = 11.84KK7 pKa = 9.45PKK9 pKa = 8.16QQRR12 pKa = 11.84KK13 pKa = 8.12PSSRR17 pKa = 11.84LNNSSSNSSSSSKK30 pKa = 9.65RR31 pKa = 11.84RR32 pKa = 11.84HH33 pKa = 4.69RR34 pKa = 11.84HH35 pKa = 5.29KK36 pKa = 10.7PMQRR40 pKa = 11.84PKK42 pKa = 10.36LQPRR46 pKa = 11.84HH47 pKa = 5.93KK48 pKa = 9.96LQPRR52 pKa = 11.84LKK54 pKa = 9.05PLHH57 pKa = 6.19RR58 pKa = 11.84LKK60 pKa = 10.56PLHH63 pKa = 6.74RR64 pKa = 11.84LKK66 pKa = 10.36QQRR69 pKa = 11.84KK70 pKa = 8.54LKK72 pKa = 8.2QQRR75 pKa = 11.84KK76 pKa = 8.37LSNRR80 pKa = 11.84LSNRR84 pKa = 11.84LSNRR88 pKa = 11.84PNNSNNSSNNNNNNNNNNNNNNNNNNSSNNSSNNSSNNSRR128 pKa = 11.84LSSNNNSSSSSNNNSSSSIKK148 pKa = 10.42YY149 pKa = 8.8NNHH152 pKa = 4.55NKK154 pKa = 9.31RR155 pKa = 11.84RR156 pKa = 11.84NRR158 pKa = 11.84SNSTNPNPISSSSSNNLSLPIIIPLSSPLLNHH190 pKa = 6.49TT191 pKa = 4.49
MM1 pKa = 6.86QRR3 pKa = 11.84PRR5 pKa = 11.84RR6 pKa = 11.84KK7 pKa = 9.45PKK9 pKa = 8.16QQRR12 pKa = 11.84KK13 pKa = 8.12PSSRR17 pKa = 11.84LNNSSSNSSSSSKK30 pKa = 9.65RR31 pKa = 11.84RR32 pKa = 11.84HH33 pKa = 4.69RR34 pKa = 11.84HH35 pKa = 5.29KK36 pKa = 10.7PMQRR40 pKa = 11.84PKK42 pKa = 10.36LQPRR46 pKa = 11.84HH47 pKa = 5.93KK48 pKa = 9.96LQPRR52 pKa = 11.84LKK54 pKa = 9.05PLHH57 pKa = 6.19RR58 pKa = 11.84LKK60 pKa = 10.56PLHH63 pKa = 6.74RR64 pKa = 11.84LKK66 pKa = 10.36QQRR69 pKa = 11.84KK70 pKa = 8.54LKK72 pKa = 8.2QQRR75 pKa = 11.84KK76 pKa = 8.37LSNRR80 pKa = 11.84LSNRR84 pKa = 11.84LSNRR88 pKa = 11.84PNNSNNSSNNNNNNNNNNNNNNNNNNSSNNSSNNSSNNSRR128 pKa = 11.84LSSNNNSSSSSNNNSSSSIKK148 pKa = 10.42YY149 pKa = 8.8NNHH152 pKa = 4.55NKK154 pKa = 9.31RR155 pKa = 11.84RR156 pKa = 11.84NRR158 pKa = 11.84SNSTNPNPISSSSSNNLSLPIIIPLSSPLLNHH190 pKa = 6.49TT191 pKa = 4.49
Molecular weight: 21.84 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2778377 |
66 |
4749 |
494.4 |
54.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.344 ± 0.037 | 1.053 ± 0.01 |
6.013 ± 0.024 | 6.392 ± 0.041 |
3.528 ± 0.021 | 6.521 ± 0.033 |
2.281 ± 0.015 | 4.919 ± 0.02 |
5.223 ± 0.034 | 8.523 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.494 ± 0.015 | 3.848 ± 0.017 |
5.703 ± 0.033 | 4.013 ± 0.032 |
5.782 ± 0.024 | 8.325 ± 0.038 |
6.016 ± 0.025 | 6.118 ± 0.021 |
1.266 ± 0.011 | 2.637 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
