
Flavobacterium sp. ALD4
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Flavobacterium; unclassified Flavobacterium
Average proteome isoelectric point is 6.59
Get precalculated fractions of proteins
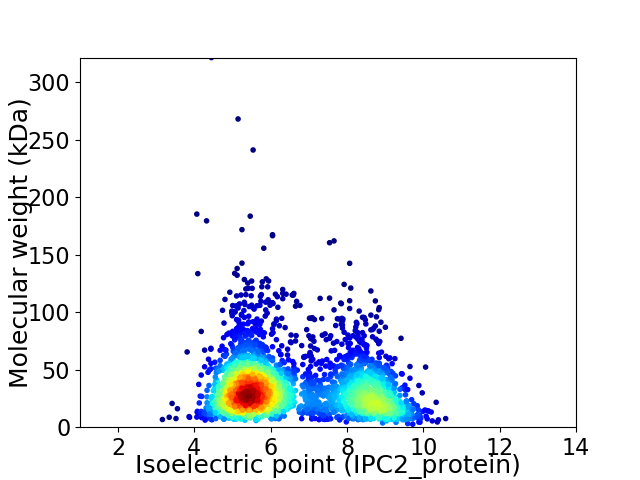
Virtual 2D-PAGE plot for 2796 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2N1G3C0|A0A2N1G3C0_9FLAO Orotidine-5'-phosphate decarboxylase OS=Flavobacterium sp. ALD4 OX=2058314 GN=pyrF PE=4 SV=1
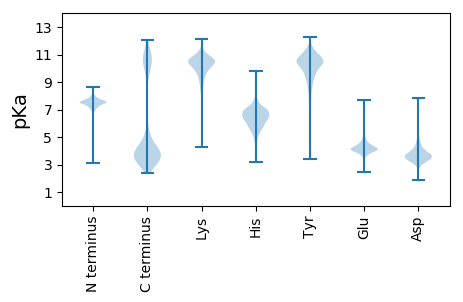
MM1 pKa = 7.0KK2 pKa = 10.21TNDD5 pKa = 3.37ILIGEE10 pKa = 4.79DD11 pKa = 3.1GCANLVVGSIVTYY24 pKa = 9.78TFTVTNPGNVSLYY37 pKa = 9.98NVTVADD43 pKa = 3.71PHH45 pKa = 7.25VGLSAIALLSGGDD58 pKa = 3.5ANTNNILEE66 pKa = 4.39VTEE69 pKa = 4.07TWTYY73 pKa = 7.65TATYY77 pKa = 8.98TVTQADD83 pKa = 3.43IDD85 pKa = 3.84EE86 pKa = 4.64GKK88 pKa = 8.39ITNQASVDD96 pKa = 3.5GFAPDD101 pKa = 3.45EE102 pKa = 4.51TKK104 pKa = 10.86VSDD107 pKa = 4.79LSGDD111 pKa = 3.71GANTNEE117 pKa = 4.16EE118 pKa = 4.11NVIPICSSPNIAIVKK133 pKa = 9.21TNDD136 pKa = 2.9ILIGEE141 pKa = 4.78DD142 pKa = 3.38GCVILALGDD151 pKa = 3.7IITYY155 pKa = 8.15TFTVTNPGNVSLHH168 pKa = 5.35NVAVQDD174 pKa = 3.75LHH176 pKa = 6.91PGLSSIALISGVDD189 pKa = 3.33ANTNNILEE197 pKa = 4.36VTEE200 pKa = 3.87TWIYY204 pKa = 9.29RR205 pKa = 11.84ATYY208 pKa = 10.55AVTQADD214 pKa = 3.26IDD216 pKa = 3.78AGKK219 pKa = 8.5ITNQASVDD227 pKa = 3.5GFAPDD232 pKa = 3.45EE233 pKa = 4.51TKK235 pKa = 10.84VSDD238 pKa = 3.87LSGSSEE244 pKa = 4.3TNNDD248 pKa = 2.97ADD250 pKa = 3.98VLIICTTPSITITKK264 pKa = 10.24DD265 pKa = 3.39GIFTDD270 pKa = 3.93STSPTGITNPGDD282 pKa = 3.62TITYY286 pKa = 7.76TFVIEE291 pKa = 4.01NTGNVTLTNVTIADD305 pKa = 4.35PLPGIVITGGPLTSMAAGASNSTTFIGTYY334 pKa = 9.72TITQADD340 pKa = 3.44INAGVVYY347 pKa = 10.76NLATVSGTQPSGTVVTATSTDD368 pKa = 3.55PTPCAICPPKK378 pKa = 9.96PDD380 pKa = 3.92CTGPDD385 pKa = 3.48CANYY389 pKa = 9.36TVTPLPNLKK398 pKa = 8.48VTKK401 pKa = 9.1TATVTTRR408 pKa = 11.84DD409 pKa = 3.88IIDD412 pKa = 3.49EE413 pKa = 4.41VYY415 pKa = 11.08SFVGDD420 pKa = 3.58VINYY424 pKa = 7.99NIRR427 pKa = 11.84VEE429 pKa = 4.07NTGYY433 pKa = 8.46ITILDD438 pKa = 3.8IVVQDD443 pKa = 4.54PLTGLNNTIATLAPGEE459 pKa = 4.14YY460 pKa = 9.68KK461 pKa = 10.68DD462 pKa = 4.5FSEE465 pKa = 4.36SHH467 pKa = 5.79TVTIVDD473 pKa = 4.4LEE475 pKa = 4.22VDD477 pKa = 3.82SVLNIATADD486 pKa = 3.59GFTQTGSPIHH496 pKa = 7.26AEE498 pKa = 3.85DD499 pKa = 3.61SLTVEE504 pKa = 4.32KK505 pKa = 10.89ASVLGCEE512 pKa = 4.86SILVHH517 pKa = 6.66NAFSPNGDD525 pKa = 4.16GINEE529 pKa = 4.04MFVIDD534 pKa = 5.13GIEE537 pKa = 3.75DD538 pKa = 4.3TICYY542 pKa = 9.49PEE544 pKa = 4.16NTVEE548 pKa = 4.81IYY550 pKa = 10.75NRR552 pKa = 11.84WGVLVFEE559 pKa = 4.49TQNYY563 pKa = 8.87NNQTNAFDD571 pKa = 4.4GYY573 pKa = 10.89SRR575 pKa = 11.84GRR577 pKa = 11.84TTIDD581 pKa = 3.17KK582 pKa = 11.04SSGLPTGTYY591 pKa = 9.94FYY593 pKa = 10.24ILKK596 pKa = 8.39YY597 pKa = 9.6TSVDD601 pKa = 3.74LNGNTQANQKK611 pKa = 10.45DD612 pKa = 3.91GYY614 pKa = 11.19LYY616 pKa = 9.48LTKK619 pKa = 10.77
MM1 pKa = 7.0KK2 pKa = 10.21TNDD5 pKa = 3.37ILIGEE10 pKa = 4.79DD11 pKa = 3.1GCANLVVGSIVTYY24 pKa = 9.78TFTVTNPGNVSLYY37 pKa = 9.98NVTVADD43 pKa = 3.71PHH45 pKa = 7.25VGLSAIALLSGGDD58 pKa = 3.5ANTNNILEE66 pKa = 4.39VTEE69 pKa = 4.07TWTYY73 pKa = 7.65TATYY77 pKa = 8.98TVTQADD83 pKa = 3.43IDD85 pKa = 3.84EE86 pKa = 4.64GKK88 pKa = 8.39ITNQASVDD96 pKa = 3.5GFAPDD101 pKa = 3.45EE102 pKa = 4.51TKK104 pKa = 10.86VSDD107 pKa = 4.79LSGDD111 pKa = 3.71GANTNEE117 pKa = 4.16EE118 pKa = 4.11NVIPICSSPNIAIVKK133 pKa = 9.21TNDD136 pKa = 2.9ILIGEE141 pKa = 4.78DD142 pKa = 3.38GCVILALGDD151 pKa = 3.7IITYY155 pKa = 8.15TFTVTNPGNVSLHH168 pKa = 5.35NVAVQDD174 pKa = 3.75LHH176 pKa = 6.91PGLSSIALISGVDD189 pKa = 3.33ANTNNILEE197 pKa = 4.36VTEE200 pKa = 3.87TWIYY204 pKa = 9.29RR205 pKa = 11.84ATYY208 pKa = 10.55AVTQADD214 pKa = 3.26IDD216 pKa = 3.78AGKK219 pKa = 8.5ITNQASVDD227 pKa = 3.5GFAPDD232 pKa = 3.45EE233 pKa = 4.51TKK235 pKa = 10.84VSDD238 pKa = 3.87LSGSSEE244 pKa = 4.3TNNDD248 pKa = 2.97ADD250 pKa = 3.98VLIICTTPSITITKK264 pKa = 10.24DD265 pKa = 3.39GIFTDD270 pKa = 3.93STSPTGITNPGDD282 pKa = 3.62TITYY286 pKa = 7.76TFVIEE291 pKa = 4.01NTGNVTLTNVTIADD305 pKa = 4.35PLPGIVITGGPLTSMAAGASNSTTFIGTYY334 pKa = 9.72TITQADD340 pKa = 3.44INAGVVYY347 pKa = 10.76NLATVSGTQPSGTVVTATSTDD368 pKa = 3.55PTPCAICPPKK378 pKa = 9.96PDD380 pKa = 3.92CTGPDD385 pKa = 3.48CANYY389 pKa = 9.36TVTPLPNLKK398 pKa = 8.48VTKK401 pKa = 9.1TATVTTRR408 pKa = 11.84DD409 pKa = 3.88IIDD412 pKa = 3.49EE413 pKa = 4.41VYY415 pKa = 11.08SFVGDD420 pKa = 3.58VINYY424 pKa = 7.99NIRR427 pKa = 11.84VEE429 pKa = 4.07NTGYY433 pKa = 8.46ITILDD438 pKa = 3.8IVVQDD443 pKa = 4.54PLTGLNNTIATLAPGEE459 pKa = 4.14YY460 pKa = 9.68KK461 pKa = 10.68DD462 pKa = 4.5FSEE465 pKa = 4.36SHH467 pKa = 5.79TVTIVDD473 pKa = 4.4LEE475 pKa = 4.22VDD477 pKa = 3.82SVLNIATADD486 pKa = 3.59GFTQTGSPIHH496 pKa = 7.26AEE498 pKa = 3.85DD499 pKa = 3.61SLTVEE504 pKa = 4.32KK505 pKa = 10.89ASVLGCEE512 pKa = 4.86SILVHH517 pKa = 6.66NAFSPNGDD525 pKa = 4.16GINEE529 pKa = 4.04MFVIDD534 pKa = 5.13GIEE537 pKa = 3.75DD538 pKa = 4.3TICYY542 pKa = 9.49PEE544 pKa = 4.16NTVEE548 pKa = 4.81IYY550 pKa = 10.75NRR552 pKa = 11.84WGVLVFEE559 pKa = 4.49TQNYY563 pKa = 8.87NNQTNAFDD571 pKa = 4.4GYY573 pKa = 10.89SRR575 pKa = 11.84GRR577 pKa = 11.84TTIDD581 pKa = 3.17KK582 pKa = 11.04SSGLPTGTYY591 pKa = 9.94FYY593 pKa = 10.24ILKK596 pKa = 8.39YY597 pKa = 9.6TSVDD601 pKa = 3.74LNGNTQANQKK611 pKa = 10.45DD612 pKa = 3.91GYY614 pKa = 11.19LYY616 pKa = 9.48LTKK619 pKa = 10.77
Molecular weight: 65.41 kDa
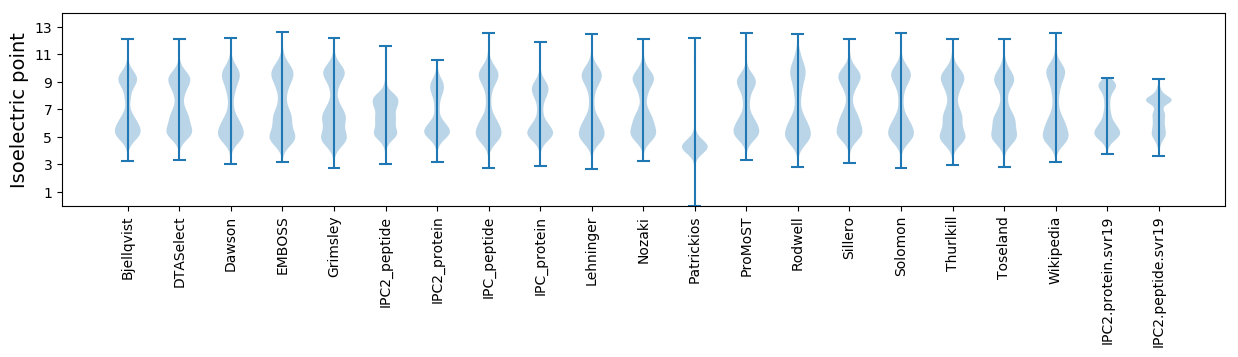
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2N1G7U7|A0A2N1G7U7_9FLAO Mannose-1-phosphate guanylyltransferase OS=Flavobacterium sp. ALD4 OX=2058314 GN=CXF59_06990 PE=4 SV=1
MM1 pKa = 7.54SVRR4 pKa = 11.84KK5 pKa = 9.68LKK7 pKa = 10.5PITPGQRR14 pKa = 11.84FRR16 pKa = 11.84VVNGYY21 pKa = 9.53DD22 pKa = 4.32AITTDD27 pKa = 3.14KK28 pKa = 10.95PEE30 pKa = 4.34RR31 pKa = 11.84SLIAPIKK38 pKa = 10.47NSGGRR43 pKa = 11.84NSQGKK48 pKa = 6.33MTMRR52 pKa = 11.84YY53 pKa = 7.39TGGGHH58 pKa = 4.86KK59 pKa = 9.44QRR61 pKa = 11.84YY62 pKa = 8.74RR63 pKa = 11.84IIDD66 pKa = 3.78FKK68 pKa = 10.31RR69 pKa = 11.84TKK71 pKa = 10.12EE72 pKa = 4.27GIPATVKK79 pKa = 10.68SIEE82 pKa = 4.0YY83 pKa = 10.15DD84 pKa = 3.43PNRR87 pKa = 11.84TAFIALLAYY96 pKa = 10.31ADD98 pKa = 3.92GEE100 pKa = 4.34KK101 pKa = 10.14TYY103 pKa = 11.26VIAQNGLKK111 pKa = 10.24VGQKK115 pKa = 10.04LLSGPEE121 pKa = 3.96SQPEE125 pKa = 3.92IGNTLPLSRR134 pKa = 11.84IPLGTVISCIEE145 pKa = 3.9LRR147 pKa = 11.84PGQGAVIARR156 pKa = 11.84SAGTFAQLMARR167 pKa = 11.84DD168 pKa = 4.06GKK170 pKa = 10.75YY171 pKa = 9.05ATIKK175 pKa = 9.6MPSGEE180 pKa = 4.18TRR182 pKa = 11.84LILLTCAATIGAVSNSDD199 pKa = 3.15HH200 pKa = 6.2QLVVSGKK207 pKa = 9.95AGRR210 pKa = 11.84TRR212 pKa = 11.84WLGRR216 pKa = 11.84RR217 pKa = 11.84PRR219 pKa = 11.84TRR221 pKa = 11.84PVAMNPVDD229 pKa = 3.63HH230 pKa = 7.13PMGGGEE236 pKa = 4.08GRR238 pKa = 11.84SSGGHH243 pKa = 4.7PRR245 pKa = 11.84SRR247 pKa = 11.84NGIPAKK253 pKa = 10.29GYY255 pKa = 7.29RR256 pKa = 11.84TRR258 pKa = 11.84SKK260 pKa = 10.85KK261 pKa = 10.4NPSNKK266 pKa = 10.11YY267 pKa = 8.41IVEE270 pKa = 4.07RR271 pKa = 11.84RR272 pKa = 11.84KK273 pKa = 10.24KK274 pKa = 10.1
MM1 pKa = 7.54SVRR4 pKa = 11.84KK5 pKa = 9.68LKK7 pKa = 10.5PITPGQRR14 pKa = 11.84FRR16 pKa = 11.84VVNGYY21 pKa = 9.53DD22 pKa = 4.32AITTDD27 pKa = 3.14KK28 pKa = 10.95PEE30 pKa = 4.34RR31 pKa = 11.84SLIAPIKK38 pKa = 10.47NSGGRR43 pKa = 11.84NSQGKK48 pKa = 6.33MTMRR52 pKa = 11.84YY53 pKa = 7.39TGGGHH58 pKa = 4.86KK59 pKa = 9.44QRR61 pKa = 11.84YY62 pKa = 8.74RR63 pKa = 11.84IIDD66 pKa = 3.78FKK68 pKa = 10.31RR69 pKa = 11.84TKK71 pKa = 10.12EE72 pKa = 4.27GIPATVKK79 pKa = 10.68SIEE82 pKa = 4.0YY83 pKa = 10.15DD84 pKa = 3.43PNRR87 pKa = 11.84TAFIALLAYY96 pKa = 10.31ADD98 pKa = 3.92GEE100 pKa = 4.34KK101 pKa = 10.14TYY103 pKa = 11.26VIAQNGLKK111 pKa = 10.24VGQKK115 pKa = 10.04LLSGPEE121 pKa = 3.96SQPEE125 pKa = 3.92IGNTLPLSRR134 pKa = 11.84IPLGTVISCIEE145 pKa = 3.9LRR147 pKa = 11.84PGQGAVIARR156 pKa = 11.84SAGTFAQLMARR167 pKa = 11.84DD168 pKa = 4.06GKK170 pKa = 10.75YY171 pKa = 9.05ATIKK175 pKa = 9.6MPSGEE180 pKa = 4.18TRR182 pKa = 11.84LILLTCAATIGAVSNSDD199 pKa = 3.15HH200 pKa = 6.2QLVVSGKK207 pKa = 9.95AGRR210 pKa = 11.84TRR212 pKa = 11.84WLGRR216 pKa = 11.84RR217 pKa = 11.84PRR219 pKa = 11.84TRR221 pKa = 11.84PVAMNPVDD229 pKa = 3.63HH230 pKa = 7.13PMGGGEE236 pKa = 4.08GRR238 pKa = 11.84SSGGHH243 pKa = 4.7PRR245 pKa = 11.84SRR247 pKa = 11.84NGIPAKK253 pKa = 10.29GYY255 pKa = 7.29RR256 pKa = 11.84TRR258 pKa = 11.84SKK260 pKa = 10.85KK261 pKa = 10.4NPSNKK266 pKa = 10.11YY267 pKa = 8.41IVEE270 pKa = 4.07RR271 pKa = 11.84RR272 pKa = 11.84KK273 pKa = 10.24KK274 pKa = 10.1
Molecular weight: 29.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
921015 |
23 |
2901 |
329.4 |
37.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.7 ± 0.045 | 0.724 ± 0.014 |
5.351 ± 0.032 | 6.361 ± 0.041 |
5.287 ± 0.037 | 6.422 ± 0.047 |
1.661 ± 0.019 | 8.304 ± 0.049 |
7.891 ± 0.044 | 9.31 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.361 ± 0.025 | 6.006 ± 0.046 |
3.315 ± 0.024 | 3.295 ± 0.023 |
3.246 ± 0.027 | 6.565 ± 0.037 |
5.866 ± 0.043 | 6.414 ± 0.04 |
0.957 ± 0.013 | 3.962 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
