
Natrinema pellirubrum (strain DSM 15624 / CIP 106293 / JCM 10476 / NCIMB 786 / 157)
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Natrialbales; Natrialbaceae; Natrinema; Natrinema pellirubrum
Average proteome isoelectric point is 4.85
Get precalculated fractions of proteins
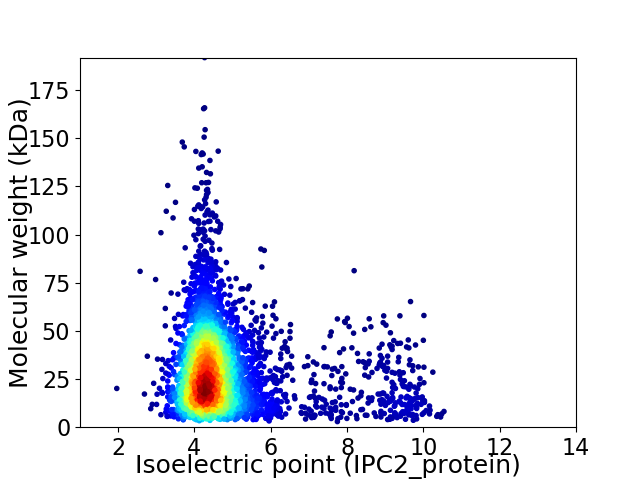
Virtual 2D-PAGE plot for 4138 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|L0JP37|L0JP37_NATP1 Short-chain dehydrogenase/reductase SDR OS=Natrinema pellirubrum (strain DSM 15624 / CIP 106293 / JCM 10476 / NCIMB 786 / 157) OX=797303 GN=Natpe_2819 PE=3 SV=1
MM1 pKa = 7.18TRR3 pKa = 11.84LTRR6 pKa = 11.84RR7 pKa = 11.84STLKK11 pKa = 10.68VAGASLAAATVPATAAAAEE30 pKa = 4.49SDD32 pKa = 4.15DD33 pKa = 3.15GWTVVEE39 pKa = 4.56TPVDD43 pKa = 3.88STLHH47 pKa = 5.97DD48 pKa = 3.61VAYY51 pKa = 8.44TATNPHH57 pKa = 6.12AVAGGGLVIEE67 pKa = 4.73RR68 pKa = 11.84TATGWEE74 pKa = 4.28VVLQGGPTGNGNDD87 pKa = 4.05LYY89 pKa = 11.45GVDD92 pKa = 3.64TTDD95 pKa = 3.47DD96 pKa = 4.38GEE98 pKa = 4.36RR99 pKa = 11.84LWIVGSSGAIGEE111 pKa = 4.31YY112 pKa = 10.65DD113 pKa = 3.62VTTGNLVDD121 pKa = 4.9RR122 pKa = 11.84SAPNDD127 pKa = 3.21YY128 pKa = 8.56TANFNDD134 pKa = 3.71VSVTGPAGEE143 pKa = 4.17ADD145 pKa = 4.12VYY147 pKa = 11.34VADD150 pKa = 4.89DD151 pKa = 4.03SGSIHH156 pKa = 6.94YY157 pKa = 10.87SFDD160 pKa = 3.3NGKK163 pKa = 9.97EE164 pKa = 4.2GTWNYY169 pKa = 8.14EE170 pKa = 4.02VPGSGSGLNAIEE182 pKa = 5.14CYY184 pKa = 10.62ADD186 pKa = 3.66GAGHH190 pKa = 7.34VIDD193 pKa = 4.17TNGKK197 pKa = 9.1VFATDD202 pKa = 5.9DD203 pKa = 3.91GVTWNPIGIEE213 pKa = 4.14DD214 pKa = 4.96AGVTYY219 pKa = 10.92YY220 pKa = 11.4GLDD223 pKa = 3.33SDD225 pKa = 5.77AEE227 pKa = 4.31DD228 pKa = 3.69DD229 pKa = 4.47VYY231 pKa = 11.91VSGGNASVFDD241 pKa = 4.05YY242 pKa = 10.64TGSRR246 pKa = 11.84WIPEE250 pKa = 4.29SLGDD254 pKa = 3.62AALFDD259 pKa = 4.35IEE261 pKa = 4.59TEE263 pKa = 3.83AGTGYY268 pKa = 8.83TVGSGGAVFEE278 pKa = 5.0LEE280 pKa = 4.37DD281 pKa = 4.44GEE283 pKa = 4.61WVQNTTPVGEE293 pKa = 4.25NLRR296 pKa = 11.84AVATGDD302 pKa = 3.02VDD304 pKa = 3.75IVVGSSGSLLEE315 pKa = 4.16RR316 pKa = 4.73
MM1 pKa = 7.18TRR3 pKa = 11.84LTRR6 pKa = 11.84RR7 pKa = 11.84STLKK11 pKa = 10.68VAGASLAAATVPATAAAAEE30 pKa = 4.49SDD32 pKa = 4.15DD33 pKa = 3.15GWTVVEE39 pKa = 4.56TPVDD43 pKa = 3.88STLHH47 pKa = 5.97DD48 pKa = 3.61VAYY51 pKa = 8.44TATNPHH57 pKa = 6.12AVAGGGLVIEE67 pKa = 4.73RR68 pKa = 11.84TATGWEE74 pKa = 4.28VVLQGGPTGNGNDD87 pKa = 4.05LYY89 pKa = 11.45GVDD92 pKa = 3.64TTDD95 pKa = 3.47DD96 pKa = 4.38GEE98 pKa = 4.36RR99 pKa = 11.84LWIVGSSGAIGEE111 pKa = 4.31YY112 pKa = 10.65DD113 pKa = 3.62VTTGNLVDD121 pKa = 4.9RR122 pKa = 11.84SAPNDD127 pKa = 3.21YY128 pKa = 8.56TANFNDD134 pKa = 3.71VSVTGPAGEE143 pKa = 4.17ADD145 pKa = 4.12VYY147 pKa = 11.34VADD150 pKa = 4.89DD151 pKa = 4.03SGSIHH156 pKa = 6.94YY157 pKa = 10.87SFDD160 pKa = 3.3NGKK163 pKa = 9.97EE164 pKa = 4.2GTWNYY169 pKa = 8.14EE170 pKa = 4.02VPGSGSGLNAIEE182 pKa = 5.14CYY184 pKa = 10.62ADD186 pKa = 3.66GAGHH190 pKa = 7.34VIDD193 pKa = 4.17TNGKK197 pKa = 9.1VFATDD202 pKa = 5.9DD203 pKa = 3.91GVTWNPIGIEE213 pKa = 4.14DD214 pKa = 4.96AGVTYY219 pKa = 10.92YY220 pKa = 11.4GLDD223 pKa = 3.33SDD225 pKa = 5.77AEE227 pKa = 4.31DD228 pKa = 3.69DD229 pKa = 4.47VYY231 pKa = 11.91VSGGNASVFDD241 pKa = 4.05YY242 pKa = 10.64TGSRR246 pKa = 11.84WIPEE250 pKa = 4.29SLGDD254 pKa = 3.62AALFDD259 pKa = 4.35IEE261 pKa = 4.59TEE263 pKa = 3.83AGTGYY268 pKa = 8.83TVGSGGAVFEE278 pKa = 5.0LEE280 pKa = 4.37DD281 pKa = 4.44GEE283 pKa = 4.61WVQNTTPVGEE293 pKa = 4.25NLRR296 pKa = 11.84AVATGDD302 pKa = 3.02VDD304 pKa = 3.75IVVGSSGSLLEE315 pKa = 4.16RR316 pKa = 4.73
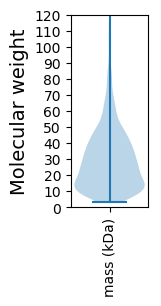
Molecular weight: 32.56 kDa
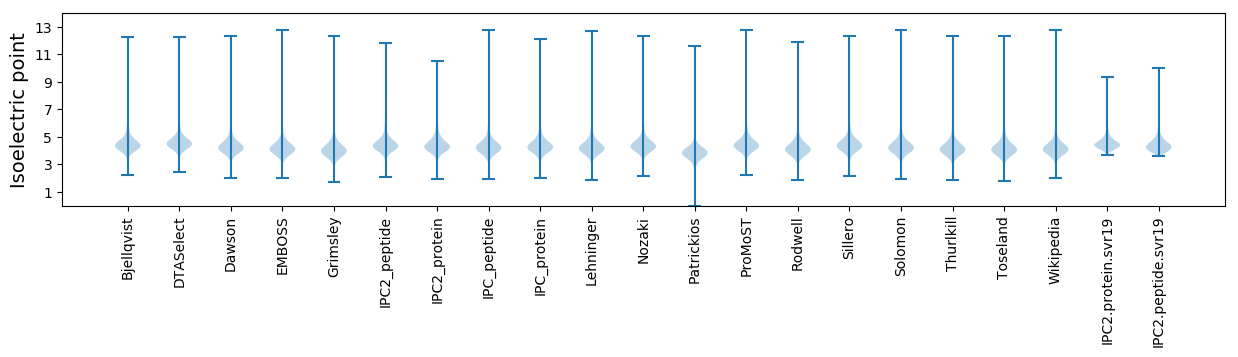
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|L0JPU9|L0JPU9_NATP1 Cobalamin B12-binding domain protein OS=Natrinema pellirubrum (strain DSM 15624 / CIP 106293 / JCM 10476 / NCIMB 786 / 157) OX=797303 GN=Natpe_2838 PE=3 SV=1
MM1 pKa = 6.85CTRR4 pKa = 11.84RR5 pKa = 11.84VRR7 pKa = 11.84RR8 pKa = 11.84SAPPLDD14 pKa = 3.73KK15 pKa = 11.05SDD17 pKa = 2.86WFHH20 pKa = 8.5RR21 pKa = 11.84MGTKK25 pKa = 9.99PGQITRR31 pKa = 11.84LLQAFLQNPSHH42 pKa = 7.08LLPVNDD48 pKa = 4.2SSPRR52 pKa = 11.84GYY54 pKa = 9.77WSPVDD59 pKa = 3.27TCRR62 pKa = 11.84RR63 pKa = 11.84FPVEE67 pKa = 3.34EE68 pKa = 4.33SEE70 pKa = 4.5PEE72 pKa = 3.48RR73 pKa = 11.84HH74 pKa = 5.3QPVVVGDD81 pKa = 4.48DD82 pKa = 2.82IDD84 pKa = 4.24RR85 pKa = 11.84PVRR88 pKa = 11.84YY89 pKa = 9.3RR90 pKa = 11.84QDD92 pKa = 2.8HH93 pKa = 6.11SRR95 pKa = 11.84TKK97 pKa = 10.43QEE99 pKa = 3.96EE100 pKa = 4.09QHH102 pKa = 6.9DD103 pKa = 4.16EE104 pKa = 3.97RR105 pKa = 11.84DD106 pKa = 4.23DD107 pKa = 3.47IRR109 pKa = 11.84EE110 pKa = 3.97TLGVHH115 pKa = 5.14TTRR118 pKa = 11.84SATLRR123 pKa = 11.84QKK125 pKa = 10.71RR126 pKa = 11.84GVDD129 pKa = 3.84PVPGSPWGHH138 pKa = 6.79PAHH141 pKa = 6.89LGHH144 pKa = 6.72SGRR147 pKa = 11.84SSDD150 pKa = 3.5PVIFGTVGLEE160 pKa = 4.18TPSKK164 pKa = 10.15RR165 pKa = 11.84FSHH168 pKa = 6.21EE169 pKa = 3.77ARR171 pKa = 11.84PGSAGVAVAIGRR183 pKa = 11.84IHH185 pKa = 6.75GLVHH189 pKa = 6.53TPPKK193 pKa = 9.86SACFRR198 pKa = 11.84LTEE201 pKa = 4.22RR202 pKa = 11.84PRR204 pKa = 11.84FSPTTTGPCGPVVQNSVSSGGSARR228 pKa = 11.84EE229 pKa = 3.57LHH231 pKa = 5.47TRR233 pKa = 11.84CRR235 pKa = 11.84DD236 pKa = 3.08SCVYY240 pKa = 10.58SGIGFWRR247 pKa = 11.84RR248 pKa = 11.84TSSGRR253 pKa = 11.84RR254 pKa = 11.84KK255 pKa = 9.8HH256 pKa = 5.9RR257 pKa = 3.63
MM1 pKa = 6.85CTRR4 pKa = 11.84RR5 pKa = 11.84VRR7 pKa = 11.84RR8 pKa = 11.84SAPPLDD14 pKa = 3.73KK15 pKa = 11.05SDD17 pKa = 2.86WFHH20 pKa = 8.5RR21 pKa = 11.84MGTKK25 pKa = 9.99PGQITRR31 pKa = 11.84LLQAFLQNPSHH42 pKa = 7.08LLPVNDD48 pKa = 4.2SSPRR52 pKa = 11.84GYY54 pKa = 9.77WSPVDD59 pKa = 3.27TCRR62 pKa = 11.84RR63 pKa = 11.84FPVEE67 pKa = 3.34EE68 pKa = 4.33SEE70 pKa = 4.5PEE72 pKa = 3.48RR73 pKa = 11.84HH74 pKa = 5.3QPVVVGDD81 pKa = 4.48DD82 pKa = 2.82IDD84 pKa = 4.24RR85 pKa = 11.84PVRR88 pKa = 11.84YY89 pKa = 9.3RR90 pKa = 11.84QDD92 pKa = 2.8HH93 pKa = 6.11SRR95 pKa = 11.84TKK97 pKa = 10.43QEE99 pKa = 3.96EE100 pKa = 4.09QHH102 pKa = 6.9DD103 pKa = 4.16EE104 pKa = 3.97RR105 pKa = 11.84DD106 pKa = 4.23DD107 pKa = 3.47IRR109 pKa = 11.84EE110 pKa = 3.97TLGVHH115 pKa = 5.14TTRR118 pKa = 11.84SATLRR123 pKa = 11.84QKK125 pKa = 10.71RR126 pKa = 11.84GVDD129 pKa = 3.84PVPGSPWGHH138 pKa = 6.79PAHH141 pKa = 6.89LGHH144 pKa = 6.72SGRR147 pKa = 11.84SSDD150 pKa = 3.5PVIFGTVGLEE160 pKa = 4.18TPSKK164 pKa = 10.15RR165 pKa = 11.84FSHH168 pKa = 6.21EE169 pKa = 3.77ARR171 pKa = 11.84PGSAGVAVAIGRR183 pKa = 11.84IHH185 pKa = 6.75GLVHH189 pKa = 6.53TPPKK193 pKa = 9.86SACFRR198 pKa = 11.84LTEE201 pKa = 4.22RR202 pKa = 11.84PRR204 pKa = 11.84FSPTTTGPCGPVVQNSVSSGGSARR228 pKa = 11.84EE229 pKa = 3.57LHH231 pKa = 5.47TRR233 pKa = 11.84CRR235 pKa = 11.84DD236 pKa = 3.08SCVYY240 pKa = 10.58SGIGFWRR247 pKa = 11.84RR248 pKa = 11.84TSSGRR253 pKa = 11.84RR254 pKa = 11.84KK255 pKa = 9.8HH256 pKa = 5.9RR257 pKa = 3.63
Molecular weight: 28.78 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1179910 |
29 |
1720 |
285.1 |
31.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.877 ± 0.06 | 0.781 ± 0.013 |
8.657 ± 0.049 | 8.869 ± 0.055 |
3.176 ± 0.025 | 8.331 ± 0.037 |
1.946 ± 0.019 | 4.353 ± 0.029 |
1.795 ± 0.026 | 8.954 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.694 ± 0.014 | 2.284 ± 0.022 |
4.672 ± 0.026 | 2.448 ± 0.027 |
6.651 ± 0.036 | 5.504 ± 0.028 |
6.531 ± 0.028 | 8.604 ± 0.039 |
1.139 ± 0.015 | 2.735 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
