
Rhodonellum psychrophilum GCM71 = DSM 17998
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Cytophagaceae; Rhodonellum; Rhodonellum psychrophilum
Average proteome isoelectric point is 6.61
Get precalculated fractions of proteins
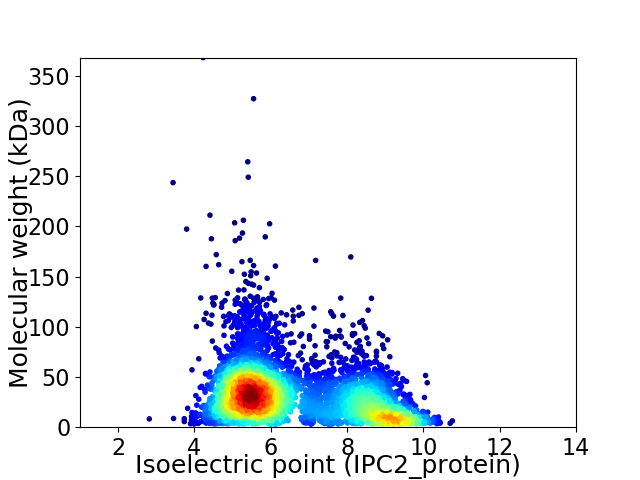
Virtual 2D-PAGE plot for 4904 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
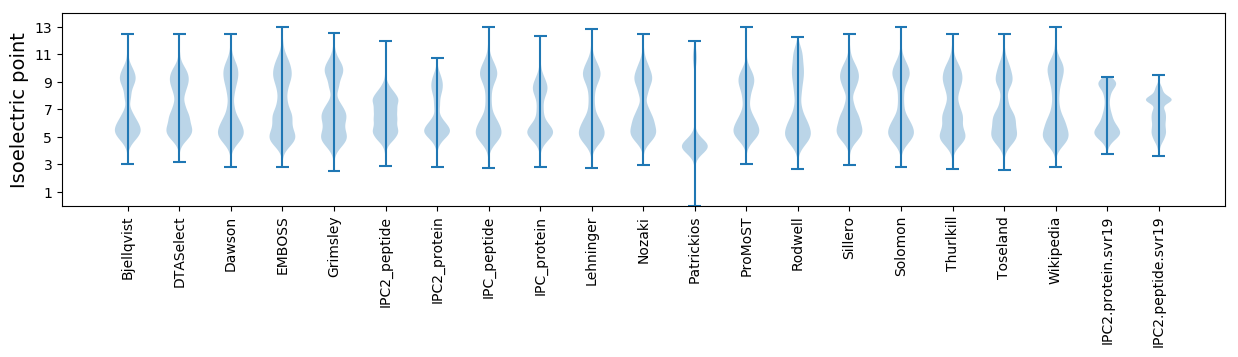
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|U5C4Q4|U5C4Q4_9BACT DUF1232 domain-containing protein OS=Rhodonellum psychrophilum GCM71 = DSM 17998 OX=1123057 GN=P872_01105 PE=4 SV=1
MM1 pKa = 7.69KK2 pKa = 10.48SPYY5 pKa = 10.71NNINFALIFVGICLISCFININKK28 pKa = 9.67SFSQGFNNNEE38 pKa = 3.78WVFGYY43 pKa = 10.39CGPNDD48 pKa = 3.88PNNYY52 pKa = 10.01LSFGKK57 pKa = 10.44SGDD60 pKa = 3.85PIVRR64 pKa = 11.84SLPGSIVVGANNNALAIDD82 pKa = 5.65PITGQKK88 pKa = 10.71LFFSNGEE95 pKa = 3.75LVYY98 pKa = 10.97NYY100 pKa = 10.83DD101 pKa = 3.71NVPLQGAPNGFNGDD115 pKa = 2.97FDD117 pKa = 3.98GRR119 pKa = 11.84QTVAIGALDD128 pKa = 3.93YY129 pKa = 11.51DD130 pKa = 4.26PDD132 pKa = 4.29GEE134 pKa = 4.25KK135 pKa = 10.65LFYY138 pKa = 10.77AFYY141 pKa = 10.28ISPSGQLLYY150 pKa = 11.07SVVDD154 pKa = 3.6MNAPGGASANQPPLGAVTALDD175 pKa = 3.79QPIGPASGAIAVVKK189 pKa = 8.91TAASPSYY196 pKa = 10.31LISFAGGNLISRR208 pKa = 11.84SIEE211 pKa = 3.83STQGDD216 pKa = 3.96FTLTATQGIPFTPKK230 pKa = 9.96AIVFNEE236 pKa = 4.14STGQLILIPEE246 pKa = 4.13NAIEE250 pKa = 5.12DD251 pKa = 4.22LLLVDD256 pKa = 5.98FDD258 pKa = 4.31TATGSFGAVTPISQSGSAEE277 pKa = 4.16EE278 pKa = 4.08IEE280 pKa = 4.57GASFSPDD287 pKa = 2.63GVFVYY292 pKa = 8.36FSRR295 pKa = 11.84GDD297 pKa = 3.31EE298 pKa = 4.11LLRR301 pKa = 11.84VPVADD306 pKa = 5.11LGATPSVIPFGNDD319 pKa = 2.06IFKK322 pKa = 11.1VYY324 pKa = 10.23DD325 pKa = 3.9LKK327 pKa = 10.91IGPDD331 pKa = 3.09GKK333 pKa = 10.75LYY335 pKa = 10.51YY336 pKa = 10.43LYY338 pKa = 10.7EE339 pKa = 4.26EE340 pKa = 4.56VLGGPQLIGVVNNPNEE356 pKa = 4.11SDD358 pKa = 3.94LDD360 pKa = 4.05LLEE363 pKa = 6.07LDD365 pKa = 4.13EE366 pKa = 6.3DD367 pKa = 4.52PFNGTDD373 pKa = 3.4FCGRR377 pKa = 11.84VFPQFAPNQDD387 pKa = 3.1INPTVDD393 pKa = 4.74FEE395 pKa = 4.91WNPEE399 pKa = 4.16EE400 pKa = 4.39PCSNNPVQLTSLITPEE416 pKa = 3.69NYY418 pKa = 9.92RR419 pKa = 11.84PVSFVWTFEE428 pKa = 3.96PALTDD433 pKa = 3.45EE434 pKa = 5.49DD435 pKa = 4.34GQPVDD440 pKa = 3.07IDD442 pKa = 3.89FNQEE446 pKa = 3.34HH447 pKa = 6.79LLIPEE452 pKa = 4.53EE453 pKa = 4.07ATADD457 pKa = 3.48QSITVTLTVTFADD470 pKa = 4.5GSVVPVTKK478 pKa = 10.32TITLKK483 pKa = 11.12EE484 pKa = 3.93NDD486 pKa = 3.55LEE488 pKa = 4.85ANFSAQDD495 pKa = 3.4TTVCEE500 pKa = 4.58GACVDD505 pKa = 3.6IGSLLEE511 pKa = 3.96VQKK514 pKa = 11.33GGGGDD519 pKa = 3.7GEE521 pKa = 4.39NPNVGGGGGGQQEE534 pKa = 4.42NYY536 pKa = 10.43EE537 pKa = 4.42YY538 pKa = 10.51FWSNRR543 pKa = 11.84RR544 pKa = 11.84GEE546 pKa = 4.28GWVKK550 pKa = 10.86DD551 pKa = 3.68KK552 pKa = 10.97EE553 pKa = 4.41NCVLLPGLYY562 pKa = 8.29WVLVRR567 pKa = 11.84EE568 pKa = 4.43EE569 pKa = 5.13GSDD572 pKa = 3.61CYY574 pKa = 10.94AYY576 pKa = 10.67ASIRR580 pKa = 11.84VKK582 pKa = 10.29IWDD585 pKa = 5.32LPDD588 pKa = 3.02QSNNVWYY595 pKa = 10.11FGDD598 pKa = 4.18GAGLDD603 pKa = 4.31FNPDD607 pKa = 3.68PNDD610 pKa = 3.43PNAPVPRR617 pKa = 11.84PVSHH621 pKa = 7.09PTNIPAGTTTISDD634 pKa = 3.26EE635 pKa = 4.2TGQVLFYY642 pKa = 10.88TDD644 pKa = 5.37GEE646 pKa = 4.97TVWDD650 pKa = 4.2LNGDD654 pKa = 3.65TMANGDD660 pKa = 4.39SIGGSNLATQSVVAVPLPQDD680 pKa = 3.08EE681 pKa = 4.94TIFYY685 pKa = 10.94LFTTQKK691 pKa = 11.15AEE693 pKa = 4.45DD694 pKa = 3.73GTNQVKK700 pKa = 10.21FSLVDD705 pKa = 3.33IKK707 pKa = 11.25VDD709 pKa = 3.31NPTGVGNVVSKK720 pKa = 11.44DD721 pKa = 3.27NFLFSPSTEE730 pKa = 3.65HH731 pKa = 7.25SAGLNSGDD739 pKa = 3.71TTWVIFHH746 pKa = 6.87EE747 pKa = 4.64LGNNTFRR754 pKa = 11.84TYY756 pKa = 10.84PVTTFGIGPPVLSSVGSAHH775 pKa = 6.09TFGSGVGSMKK785 pKa = 10.49FSPDD789 pKa = 2.99GSKK792 pKa = 10.73LAVTIQNGSCSRR804 pKa = 11.84LEE806 pKa = 3.75IFDD809 pKa = 4.55FNQTTGKK816 pKa = 8.1LTEE819 pKa = 4.07YY820 pKa = 11.22ALLEE824 pKa = 4.17LSCSGEE830 pKa = 3.92DD831 pKa = 3.61VYY833 pKa = 11.69GLEE836 pKa = 5.53FSDD839 pKa = 3.72DD840 pKa = 3.73SNRR843 pKa = 11.84VFVSYY848 pKa = 10.11TGGGGKK854 pKa = 8.85IEE856 pKa = 4.27EE857 pKa = 4.25FLIKK861 pKa = 10.94NPDD864 pKa = 3.31AQGAATPSACATCFEE879 pKa = 4.33NAGDD883 pKa = 3.74KK884 pKa = 10.94AAVEE888 pKa = 4.36SCILGTRR895 pKa = 11.84NTLSSAGPFGALQIGPDD912 pKa = 3.55GQIYY916 pKa = 8.59VARR919 pKa = 11.84PGQNVLGTINAGQGCANSAYY939 pKa = 10.72VEE941 pKa = 4.4MGTSTLTGTTNLGLPSFVQNSGSSIPEE968 pKa = 3.9PTLGGPDD975 pKa = 3.94RR976 pKa = 11.84LCLDD980 pKa = 4.05PEE982 pKa = 4.61NGTEE986 pKa = 4.07ALFEE990 pKa = 4.78GGGEE994 pKa = 4.04PDD996 pKa = 2.93IDD998 pKa = 3.99SYY1000 pKa = 11.65FWTIVSEE1007 pKa = 4.0NGQAVLTNFGGPGEE1021 pKa = 4.12EE1022 pKa = 4.4FQTLTHH1028 pKa = 6.38VFQEE1032 pKa = 4.7DD1033 pKa = 4.14GTFTVSLRR1041 pKa = 11.84VDD1043 pKa = 3.03RR1044 pKa = 11.84CGDD1047 pKa = 3.27ANYY1050 pKa = 10.31FEE1052 pKa = 6.26ASMEE1056 pKa = 4.38VVVVAPPPITLVDD1069 pKa = 4.11DD1070 pKa = 3.71VTLCSGTPVTLVAIDD1085 pKa = 4.76GYY1087 pKa = 11.29DD1088 pKa = 3.73PNEE1091 pKa = 3.89GLYY1094 pKa = 10.92DD1095 pKa = 3.87FEE1097 pKa = 4.43WRR1099 pKa = 11.84NAAGQLIGDD1108 pKa = 4.21EE1109 pKa = 4.4NSNSIAVEE1117 pKa = 3.91EE1118 pKa = 4.05EE1119 pKa = 3.92SIYY1122 pKa = 10.76TVTVSYY1128 pKa = 7.6RR1129 pKa = 11.84TPSGSDD1135 pKa = 2.81TVFFDD1140 pKa = 3.25VCPASKK1146 pKa = 10.53SVFVGPAFEE1155 pKa = 5.32FDD1157 pKa = 3.38LTQSAEE1163 pKa = 4.18EE1164 pKa = 4.07VCYY1167 pKa = 11.06DD1168 pKa = 3.28EE1169 pKa = 4.53TLVVFAPNTPVIGDD1183 pKa = 3.91WYY1185 pKa = 7.9YY1186 pKa = 10.82QQVGSPTRR1194 pKa = 11.84TLIGEE1199 pKa = 4.34FFEE1202 pKa = 5.96LEE1204 pKa = 4.9LIPTDD1209 pKa = 4.13LPAPGLYY1216 pKa = 10.06EE1217 pKa = 4.29IIFITEE1223 pKa = 3.89DD1224 pKa = 4.35PIVPGCLVEE1233 pKa = 5.44KK1234 pKa = 10.53KK1235 pKa = 10.74LEE1237 pKa = 4.19LLVHH1241 pKa = 6.46PLPNFEE1247 pKa = 5.53LLVLTDD1253 pKa = 5.08ADD1255 pKa = 4.69DD1256 pKa = 4.23CATPNGSFEE1265 pKa = 3.9ITALLDD1271 pKa = 3.31AEE1273 pKa = 4.52RR1274 pKa = 11.84IEE1276 pKa = 3.99ILEE1279 pKa = 4.17TGEE1282 pKa = 4.0VFNNVNAGDD1291 pKa = 4.06VLPVISNLEE1300 pKa = 3.83PGIYY1304 pKa = 8.88TVQATNSFGCVFTRR1318 pKa = 11.84SVTIANLNPPVALEE1332 pKa = 3.85YY1333 pKa = 10.06TVAVNPEE1340 pKa = 3.73VCGPNEE1346 pKa = 4.04VEE1348 pKa = 4.04NGAVIITFSNGPQSGSYY1365 pKa = 10.71VLTRR1369 pKa = 11.84QQDD1372 pKa = 3.66GLQFSGTITNQEE1384 pKa = 3.72IVTISVPEE1392 pKa = 3.68GDD1394 pKa = 4.01YY1395 pKa = 11.19AVEE1398 pKa = 3.99VSDD1401 pKa = 5.99ALGCTLPEE1409 pKa = 4.0PVNYY1413 pKa = 10.39LVDD1416 pKa = 3.76EE1417 pKa = 4.38KK1418 pKa = 11.48LEE1420 pKa = 4.03VDD1422 pKa = 4.18FTVPTNLIACGLFSFIPQTNQSLVFTLTDD1451 pKa = 3.34ALGNGIAADD1460 pKa = 3.74ANGEE1464 pKa = 4.17FTLLASGAYY1473 pKa = 9.49QITGEE1478 pKa = 4.72DD1479 pKa = 3.91PTGIDD1484 pKa = 3.79CPRR1487 pKa = 11.84VRR1489 pKa = 11.84TMNVEE1494 pKa = 3.69INGPIGFSLSGPADD1508 pKa = 3.53PCATAISYY1516 pKa = 9.99DD1517 pKa = 3.87AILNGTDD1524 pKa = 3.32PADD1527 pKa = 4.05VFFFWKK1533 pKa = 10.16NEE1535 pKa = 3.55SGAIVGRR1542 pKa = 11.84NQTFFPAAAGEE1553 pKa = 4.19YY1554 pKa = 9.7SLEE1557 pKa = 4.08VQPRR1561 pKa = 11.84AGSLCPAAPIDD1572 pKa = 3.79FTVEE1576 pKa = 4.66DD1577 pKa = 4.66FGQPVNVVLDD1587 pKa = 3.99ALPFCGDD1594 pKa = 4.08DD1595 pKa = 3.44PFTTITVVADD1605 pKa = 3.68LTNVASIQWFSLQGGVATLLPGFDD1629 pKa = 4.16DD1630 pKa = 3.63QQTITVIEE1638 pKa = 4.2NGVYY1642 pKa = 9.85QAVLFSGFGCEE1653 pKa = 5.02LGRR1656 pKa = 11.84QQVQITKK1663 pKa = 10.58SFITPPVLEE1672 pKa = 4.16TRR1674 pKa = 11.84YY1675 pKa = 10.0VICAAEE1681 pKa = 4.05NVVNVLDD1688 pKa = 4.13PGVYY1692 pKa = 10.26DD1693 pKa = 3.5NYY1695 pKa = 10.53SWEE1698 pKa = 4.22LEE1700 pKa = 4.0GEE1702 pKa = 4.35EE1703 pKa = 4.53VSNSPTFTPTLPGNYY1718 pKa = 9.51ALTVSDD1724 pKa = 5.19EE1725 pKa = 4.22FDD1727 pKa = 3.53CLFTIEE1733 pKa = 4.76FVVEE1737 pKa = 4.08EE1738 pKa = 4.23DD1739 pKa = 4.31CEE1741 pKa = 4.3LKK1743 pKa = 10.56IIFPNAMRR1751 pKa = 11.84PSDD1754 pKa = 3.9PSRR1757 pKa = 11.84LFVVYY1762 pKa = 10.7ANDD1765 pKa = 4.52FIDD1768 pKa = 4.31HH1769 pKa = 6.34VDD1771 pKa = 3.35VLIYY1775 pKa = 10.61NRR1777 pKa = 11.84WGEE1780 pKa = 4.16LIYY1783 pKa = 10.7FCEE1786 pKa = 4.33SEE1788 pKa = 4.2NVQGEE1793 pKa = 4.47EE1794 pKa = 5.18VLCTWDD1800 pKa = 3.35GTVGGRR1806 pKa = 11.84VVPIGTYY1813 pKa = 9.11PVIVRR1818 pKa = 11.84FRR1820 pKa = 11.84SEE1822 pKa = 3.57NQNIVKK1828 pKa = 8.94TIRR1831 pKa = 11.84KK1832 pKa = 8.59SIVVIEE1838 pKa = 4.1
MM1 pKa = 7.69KK2 pKa = 10.48SPYY5 pKa = 10.71NNINFALIFVGICLISCFININKK28 pKa = 9.67SFSQGFNNNEE38 pKa = 3.78WVFGYY43 pKa = 10.39CGPNDD48 pKa = 3.88PNNYY52 pKa = 10.01LSFGKK57 pKa = 10.44SGDD60 pKa = 3.85PIVRR64 pKa = 11.84SLPGSIVVGANNNALAIDD82 pKa = 5.65PITGQKK88 pKa = 10.71LFFSNGEE95 pKa = 3.75LVYY98 pKa = 10.97NYY100 pKa = 10.83DD101 pKa = 3.71NVPLQGAPNGFNGDD115 pKa = 2.97FDD117 pKa = 3.98GRR119 pKa = 11.84QTVAIGALDD128 pKa = 3.93YY129 pKa = 11.51DD130 pKa = 4.26PDD132 pKa = 4.29GEE134 pKa = 4.25KK135 pKa = 10.65LFYY138 pKa = 10.77AFYY141 pKa = 10.28ISPSGQLLYY150 pKa = 11.07SVVDD154 pKa = 3.6MNAPGGASANQPPLGAVTALDD175 pKa = 3.79QPIGPASGAIAVVKK189 pKa = 8.91TAASPSYY196 pKa = 10.31LISFAGGNLISRR208 pKa = 11.84SIEE211 pKa = 3.83STQGDD216 pKa = 3.96FTLTATQGIPFTPKK230 pKa = 9.96AIVFNEE236 pKa = 4.14STGQLILIPEE246 pKa = 4.13NAIEE250 pKa = 5.12DD251 pKa = 4.22LLLVDD256 pKa = 5.98FDD258 pKa = 4.31TATGSFGAVTPISQSGSAEE277 pKa = 4.16EE278 pKa = 4.08IEE280 pKa = 4.57GASFSPDD287 pKa = 2.63GVFVYY292 pKa = 8.36FSRR295 pKa = 11.84GDD297 pKa = 3.31EE298 pKa = 4.11LLRR301 pKa = 11.84VPVADD306 pKa = 5.11LGATPSVIPFGNDD319 pKa = 2.06IFKK322 pKa = 11.1VYY324 pKa = 10.23DD325 pKa = 3.9LKK327 pKa = 10.91IGPDD331 pKa = 3.09GKK333 pKa = 10.75LYY335 pKa = 10.51YY336 pKa = 10.43LYY338 pKa = 10.7EE339 pKa = 4.26EE340 pKa = 4.56VLGGPQLIGVVNNPNEE356 pKa = 4.11SDD358 pKa = 3.94LDD360 pKa = 4.05LLEE363 pKa = 6.07LDD365 pKa = 4.13EE366 pKa = 6.3DD367 pKa = 4.52PFNGTDD373 pKa = 3.4FCGRR377 pKa = 11.84VFPQFAPNQDD387 pKa = 3.1INPTVDD393 pKa = 4.74FEE395 pKa = 4.91WNPEE399 pKa = 4.16EE400 pKa = 4.39PCSNNPVQLTSLITPEE416 pKa = 3.69NYY418 pKa = 9.92RR419 pKa = 11.84PVSFVWTFEE428 pKa = 3.96PALTDD433 pKa = 3.45EE434 pKa = 5.49DD435 pKa = 4.34GQPVDD440 pKa = 3.07IDD442 pKa = 3.89FNQEE446 pKa = 3.34HH447 pKa = 6.79LLIPEE452 pKa = 4.53EE453 pKa = 4.07ATADD457 pKa = 3.48QSITVTLTVTFADD470 pKa = 4.5GSVVPVTKK478 pKa = 10.32TITLKK483 pKa = 11.12EE484 pKa = 3.93NDD486 pKa = 3.55LEE488 pKa = 4.85ANFSAQDD495 pKa = 3.4TTVCEE500 pKa = 4.58GACVDD505 pKa = 3.6IGSLLEE511 pKa = 3.96VQKK514 pKa = 11.33GGGGDD519 pKa = 3.7GEE521 pKa = 4.39NPNVGGGGGGQQEE534 pKa = 4.42NYY536 pKa = 10.43EE537 pKa = 4.42YY538 pKa = 10.51FWSNRR543 pKa = 11.84RR544 pKa = 11.84GEE546 pKa = 4.28GWVKK550 pKa = 10.86DD551 pKa = 3.68KK552 pKa = 10.97EE553 pKa = 4.41NCVLLPGLYY562 pKa = 8.29WVLVRR567 pKa = 11.84EE568 pKa = 4.43EE569 pKa = 5.13GSDD572 pKa = 3.61CYY574 pKa = 10.94AYY576 pKa = 10.67ASIRR580 pKa = 11.84VKK582 pKa = 10.29IWDD585 pKa = 5.32LPDD588 pKa = 3.02QSNNVWYY595 pKa = 10.11FGDD598 pKa = 4.18GAGLDD603 pKa = 4.31FNPDD607 pKa = 3.68PNDD610 pKa = 3.43PNAPVPRR617 pKa = 11.84PVSHH621 pKa = 7.09PTNIPAGTTTISDD634 pKa = 3.26EE635 pKa = 4.2TGQVLFYY642 pKa = 10.88TDD644 pKa = 5.37GEE646 pKa = 4.97TVWDD650 pKa = 4.2LNGDD654 pKa = 3.65TMANGDD660 pKa = 4.39SIGGSNLATQSVVAVPLPQDD680 pKa = 3.08EE681 pKa = 4.94TIFYY685 pKa = 10.94LFTTQKK691 pKa = 11.15AEE693 pKa = 4.45DD694 pKa = 3.73GTNQVKK700 pKa = 10.21FSLVDD705 pKa = 3.33IKK707 pKa = 11.25VDD709 pKa = 3.31NPTGVGNVVSKK720 pKa = 11.44DD721 pKa = 3.27NFLFSPSTEE730 pKa = 3.65HH731 pKa = 7.25SAGLNSGDD739 pKa = 3.71TTWVIFHH746 pKa = 6.87EE747 pKa = 4.64LGNNTFRR754 pKa = 11.84TYY756 pKa = 10.84PVTTFGIGPPVLSSVGSAHH775 pKa = 6.09TFGSGVGSMKK785 pKa = 10.49FSPDD789 pKa = 2.99GSKK792 pKa = 10.73LAVTIQNGSCSRR804 pKa = 11.84LEE806 pKa = 3.75IFDD809 pKa = 4.55FNQTTGKK816 pKa = 8.1LTEE819 pKa = 4.07YY820 pKa = 11.22ALLEE824 pKa = 4.17LSCSGEE830 pKa = 3.92DD831 pKa = 3.61VYY833 pKa = 11.69GLEE836 pKa = 5.53FSDD839 pKa = 3.72DD840 pKa = 3.73SNRR843 pKa = 11.84VFVSYY848 pKa = 10.11TGGGGKK854 pKa = 8.85IEE856 pKa = 4.27EE857 pKa = 4.25FLIKK861 pKa = 10.94NPDD864 pKa = 3.31AQGAATPSACATCFEE879 pKa = 4.33NAGDD883 pKa = 3.74KK884 pKa = 10.94AAVEE888 pKa = 4.36SCILGTRR895 pKa = 11.84NTLSSAGPFGALQIGPDD912 pKa = 3.55GQIYY916 pKa = 8.59VARR919 pKa = 11.84PGQNVLGTINAGQGCANSAYY939 pKa = 10.72VEE941 pKa = 4.4MGTSTLTGTTNLGLPSFVQNSGSSIPEE968 pKa = 3.9PTLGGPDD975 pKa = 3.94RR976 pKa = 11.84LCLDD980 pKa = 4.05PEE982 pKa = 4.61NGTEE986 pKa = 4.07ALFEE990 pKa = 4.78GGGEE994 pKa = 4.04PDD996 pKa = 2.93IDD998 pKa = 3.99SYY1000 pKa = 11.65FWTIVSEE1007 pKa = 4.0NGQAVLTNFGGPGEE1021 pKa = 4.12EE1022 pKa = 4.4FQTLTHH1028 pKa = 6.38VFQEE1032 pKa = 4.7DD1033 pKa = 4.14GTFTVSLRR1041 pKa = 11.84VDD1043 pKa = 3.03RR1044 pKa = 11.84CGDD1047 pKa = 3.27ANYY1050 pKa = 10.31FEE1052 pKa = 6.26ASMEE1056 pKa = 4.38VVVVAPPPITLVDD1069 pKa = 4.11DD1070 pKa = 3.71VTLCSGTPVTLVAIDD1085 pKa = 4.76GYY1087 pKa = 11.29DD1088 pKa = 3.73PNEE1091 pKa = 3.89GLYY1094 pKa = 10.92DD1095 pKa = 3.87FEE1097 pKa = 4.43WRR1099 pKa = 11.84NAAGQLIGDD1108 pKa = 4.21EE1109 pKa = 4.4NSNSIAVEE1117 pKa = 3.91EE1118 pKa = 4.05EE1119 pKa = 3.92SIYY1122 pKa = 10.76TVTVSYY1128 pKa = 7.6RR1129 pKa = 11.84TPSGSDD1135 pKa = 2.81TVFFDD1140 pKa = 3.25VCPASKK1146 pKa = 10.53SVFVGPAFEE1155 pKa = 5.32FDD1157 pKa = 3.38LTQSAEE1163 pKa = 4.18EE1164 pKa = 4.07VCYY1167 pKa = 11.06DD1168 pKa = 3.28EE1169 pKa = 4.53TLVVFAPNTPVIGDD1183 pKa = 3.91WYY1185 pKa = 7.9YY1186 pKa = 10.82QQVGSPTRR1194 pKa = 11.84TLIGEE1199 pKa = 4.34FFEE1202 pKa = 5.96LEE1204 pKa = 4.9LIPTDD1209 pKa = 4.13LPAPGLYY1216 pKa = 10.06EE1217 pKa = 4.29IIFITEE1223 pKa = 3.89DD1224 pKa = 4.35PIVPGCLVEE1233 pKa = 5.44KK1234 pKa = 10.53KK1235 pKa = 10.74LEE1237 pKa = 4.19LLVHH1241 pKa = 6.46PLPNFEE1247 pKa = 5.53LLVLTDD1253 pKa = 5.08ADD1255 pKa = 4.69DD1256 pKa = 4.23CATPNGSFEE1265 pKa = 3.9ITALLDD1271 pKa = 3.31AEE1273 pKa = 4.52RR1274 pKa = 11.84IEE1276 pKa = 3.99ILEE1279 pKa = 4.17TGEE1282 pKa = 4.0VFNNVNAGDD1291 pKa = 4.06VLPVISNLEE1300 pKa = 3.83PGIYY1304 pKa = 8.88TVQATNSFGCVFTRR1318 pKa = 11.84SVTIANLNPPVALEE1332 pKa = 3.85YY1333 pKa = 10.06TVAVNPEE1340 pKa = 3.73VCGPNEE1346 pKa = 4.04VEE1348 pKa = 4.04NGAVIITFSNGPQSGSYY1365 pKa = 10.71VLTRR1369 pKa = 11.84QQDD1372 pKa = 3.66GLQFSGTITNQEE1384 pKa = 3.72IVTISVPEE1392 pKa = 3.68GDD1394 pKa = 4.01YY1395 pKa = 11.19AVEE1398 pKa = 3.99VSDD1401 pKa = 5.99ALGCTLPEE1409 pKa = 4.0PVNYY1413 pKa = 10.39LVDD1416 pKa = 3.76EE1417 pKa = 4.38KK1418 pKa = 11.48LEE1420 pKa = 4.03VDD1422 pKa = 4.18FTVPTNLIACGLFSFIPQTNQSLVFTLTDD1451 pKa = 3.34ALGNGIAADD1460 pKa = 3.74ANGEE1464 pKa = 4.17FTLLASGAYY1473 pKa = 9.49QITGEE1478 pKa = 4.72DD1479 pKa = 3.91PTGIDD1484 pKa = 3.79CPRR1487 pKa = 11.84VRR1489 pKa = 11.84TMNVEE1494 pKa = 3.69INGPIGFSLSGPADD1508 pKa = 3.53PCATAISYY1516 pKa = 9.99DD1517 pKa = 3.87AILNGTDD1524 pKa = 3.32PADD1527 pKa = 4.05VFFFWKK1533 pKa = 10.16NEE1535 pKa = 3.55SGAIVGRR1542 pKa = 11.84NQTFFPAAAGEE1553 pKa = 4.19YY1554 pKa = 9.7SLEE1557 pKa = 4.08VQPRR1561 pKa = 11.84AGSLCPAAPIDD1572 pKa = 3.79FTVEE1576 pKa = 4.66DD1577 pKa = 4.66FGQPVNVVLDD1587 pKa = 3.99ALPFCGDD1594 pKa = 4.08DD1595 pKa = 3.44PFTTITVVADD1605 pKa = 3.68LTNVASIQWFSLQGGVATLLPGFDD1629 pKa = 4.16DD1630 pKa = 3.63QQTITVIEE1638 pKa = 4.2NGVYY1642 pKa = 9.85QAVLFSGFGCEE1653 pKa = 5.02LGRR1656 pKa = 11.84QQVQITKK1663 pKa = 10.58SFITPPVLEE1672 pKa = 4.16TRR1674 pKa = 11.84YY1675 pKa = 10.0VICAAEE1681 pKa = 4.05NVVNVLDD1688 pKa = 4.13PGVYY1692 pKa = 10.26DD1693 pKa = 3.5NYY1695 pKa = 10.53SWEE1698 pKa = 4.22LEE1700 pKa = 4.0GEE1702 pKa = 4.35EE1703 pKa = 4.53VSNSPTFTPTLPGNYY1718 pKa = 9.51ALTVSDD1724 pKa = 5.19EE1725 pKa = 4.22FDD1727 pKa = 3.53CLFTIEE1733 pKa = 4.76FVVEE1737 pKa = 4.08EE1738 pKa = 4.23DD1739 pKa = 4.31CEE1741 pKa = 4.3LKK1743 pKa = 10.56IIFPNAMRR1751 pKa = 11.84PSDD1754 pKa = 3.9PSRR1757 pKa = 11.84LFVVYY1762 pKa = 10.7ANDD1765 pKa = 4.52FIDD1768 pKa = 4.31HH1769 pKa = 6.34VDD1771 pKa = 3.35VLIYY1775 pKa = 10.61NRR1777 pKa = 11.84WGEE1780 pKa = 4.16LIYY1783 pKa = 10.7FCEE1786 pKa = 4.33SEE1788 pKa = 4.2NVQGEE1793 pKa = 4.47EE1794 pKa = 5.18VLCTWDD1800 pKa = 3.35GTVGGRR1806 pKa = 11.84VVPIGTYY1813 pKa = 9.11PVIVRR1818 pKa = 11.84FRR1820 pKa = 11.84SEE1822 pKa = 3.57NQNIVKK1828 pKa = 8.94TIRR1831 pKa = 11.84KK1832 pKa = 8.59SIVVIEE1838 pKa = 4.1
Molecular weight: 197.23 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|U5C5E6|U5C5E6_9BACT 50S ribosomal protein L9 OS=Rhodonellum psychrophilum GCM71 = DSM 17998 OX=1123057 GN=rplI PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84RR10 pKa = 11.84KK11 pKa = 9.62RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.5HH16 pKa = 3.99GFRR19 pKa = 11.84EE20 pKa = 4.18RR21 pKa = 11.84MSTANGRR28 pKa = 11.84RR29 pKa = 11.84VIKK32 pKa = 10.36ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.08GRR39 pKa = 11.84HH40 pKa = 5.24KK41 pKa = 10.87LSVSSEE47 pKa = 3.97KK48 pKa = 9.91TLKK51 pKa = 10.51KK52 pKa = 10.72
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84RR10 pKa = 11.84KK11 pKa = 9.62RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.5HH16 pKa = 3.99GFRR19 pKa = 11.84EE20 pKa = 4.18RR21 pKa = 11.84MSTANGRR28 pKa = 11.84RR29 pKa = 11.84VIKK32 pKa = 10.36ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.08GRR39 pKa = 11.84HH40 pKa = 5.24KK41 pKa = 10.87LSVSSEE47 pKa = 3.97KK48 pKa = 9.91TLKK51 pKa = 10.51KK52 pKa = 10.72
Molecular weight: 6.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1499400 |
29 |
3651 |
305.8 |
34.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.442 ± 0.037 | 0.67 ± 0.012 |
5.222 ± 0.022 | 6.733 ± 0.034 |
5.667 ± 0.033 | 7.143 ± 0.041 |
1.845 ± 0.019 | 7.545 ± 0.038 |
7.123 ± 0.043 | 9.816 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.527 ± 0.018 | 5.396 ± 0.034 |
3.893 ± 0.024 | 3.601 ± 0.019 |
3.926 ± 0.024 | 6.545 ± 0.03 |
5.03 ± 0.037 | 6.075 ± 0.027 |
1.219 ± 0.016 | 3.586 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
