
Lagenorhynchus acutus papillomavirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Upsilonpapillomavirus; Upsilonpapillomavirus 1; Tursiops truncatus papillomavirus 1; Tursiops truncatus papillomavirus type 3
Average proteome isoelectric point is 6.14
Get precalculated fractions of proteins
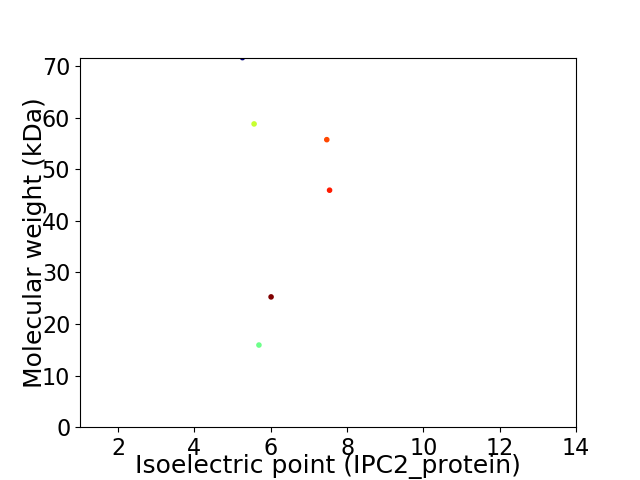
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F2VIS6|F2VIS6_9PAPI Regulatory protein E2 OS=Lagenorhynchus acutus papillomavirus OX=706530 GN=E2 PE=3 SV=1
MM1 pKa = 7.49DD2 pKa = 5.06TSPGTDD8 pKa = 3.51PLEE11 pKa = 4.7GGCNDD16 pKa = 3.24WVLLEE21 pKa = 4.11AAEE24 pKa = 4.54AGGSGIDD31 pKa = 3.57EE32 pKa = 4.39EE33 pKa = 4.44EE34 pKa = 5.66GEE36 pKa = 4.82DD37 pKa = 4.03EE38 pKa = 4.94DD39 pKa = 6.69DD40 pKa = 4.54SGEE43 pKa = 4.39DD44 pKa = 3.24MLDD47 pKa = 4.33FIDD50 pKa = 6.17DD51 pKa = 3.45SSTFQNAEE59 pKa = 3.78TQLYY63 pKa = 9.34CRR65 pKa = 11.84QLQVEE70 pKa = 4.24QQRR73 pKa = 11.84IDD75 pKa = 3.71DD76 pKa = 3.98EE77 pKa = 4.87RR78 pKa = 11.84VAQALKK84 pKa = 10.61RR85 pKa = 11.84KK86 pKa = 9.29FLDD89 pKa = 3.61SPKK92 pKa = 10.75SKK94 pKa = 10.57VDD96 pKa = 3.54SEE98 pKa = 4.24LSPRR102 pKa = 11.84LAAISLQEE110 pKa = 3.72QQRR113 pKa = 11.84RR114 pKa = 11.84GRR116 pKa = 11.84ARR118 pKa = 11.84RR119 pKa = 11.84RR120 pKa = 11.84LYY122 pKa = 10.58KK123 pKa = 10.64DD124 pKa = 2.99SGHH127 pKa = 6.54GDD129 pKa = 3.36SLEE132 pKa = 4.38AEE134 pKa = 4.57SEE136 pKa = 3.99EE137 pKa = 5.23SMVCRR142 pKa = 11.84NVQVPKK148 pKa = 10.11NQTMDD153 pKa = 3.21AVGAGEE159 pKa = 4.02GARR162 pKa = 11.84PKK164 pKa = 10.57EE165 pKa = 4.12VGNASTQEE173 pKa = 4.08VQCAGMSEE181 pKa = 4.42EE182 pKa = 4.53SACSVTQILRR192 pKa = 11.84AGKK195 pKa = 9.04PRR197 pKa = 11.84ALLLGVFKK205 pKa = 10.63EE206 pKa = 4.3AFGCSFADD214 pKa = 3.49LTRR217 pKa = 11.84CFKK220 pKa = 10.71SDD222 pKa = 3.09KK223 pKa = 9.94TVNDD227 pKa = 3.65DD228 pKa = 3.31WTCFIVGVPCSLEE241 pKa = 3.97GAISDD246 pKa = 4.06LLKK249 pKa = 10.58PSTLFTHH256 pKa = 5.85VTTATSKK263 pKa = 10.97YY264 pKa = 9.42GLTVLLLVQWKK275 pKa = 7.96TGKK278 pKa = 10.03SRR280 pKa = 11.84EE281 pKa = 4.38TVGNTLAGLLSVDD294 pKa = 3.48KK295 pKa = 10.75QQIVCEE301 pKa = 4.12PPKK304 pKa = 10.15IRR306 pKa = 11.84HH307 pKa = 6.0LGAAMFWYY315 pKa = 10.17KK316 pKa = 10.57KK317 pKa = 9.88AISSACVVTGEE328 pKa = 4.13MPVWILKK335 pKa = 9.21QVSIQDD341 pKa = 3.51QLGDD345 pKa = 3.43VCQFSLSVMIQWAYY359 pKa = 11.29DD360 pKa = 3.46NGHH363 pKa = 6.33DD364 pKa = 3.83TEE366 pKa = 4.17EE367 pKa = 5.15AIAYY371 pKa = 7.49EE372 pKa = 4.3YY373 pKa = 11.18AALADD378 pKa = 3.76EE379 pKa = 5.25DD380 pKa = 4.38KK381 pKa = 11.21NAEE384 pKa = 3.76AFLRR388 pKa = 11.84SNCQPKK394 pKa = 9.82YY395 pKa = 11.15VKK397 pKa = 10.49DD398 pKa = 3.74CANMLRR404 pKa = 11.84LYY406 pKa = 10.52KK407 pKa = 9.99RR408 pKa = 11.84AEE410 pKa = 3.82MRR412 pKa = 11.84KK413 pKa = 8.42MSMGQWIKK421 pKa = 10.58HH422 pKa = 5.65KK423 pKa = 10.28CDD425 pKa = 3.22AVQGEE430 pKa = 4.77GDD432 pKa = 3.62WKK434 pKa = 10.9HH435 pKa = 5.91VMNFLKK441 pKa = 10.72FQGIEE446 pKa = 3.78IMPFLIDD453 pKa = 4.22FRR455 pKa = 11.84LFLKK459 pKa = 10.08GIPKK463 pKa = 9.5RR464 pKa = 11.84NCMVLYY470 pKa = 10.16GPPNTGKK477 pKa = 10.5SLFAMSLIHH486 pKa = 7.02LLGGKK491 pKa = 8.88VLSYY495 pKa = 11.26VNSSSHH501 pKa = 6.44FWLQPVADD509 pKa = 4.63CKK511 pKa = 11.08VALIDD516 pKa = 5.26DD517 pKa = 4.58ATASTWDD524 pKa = 3.48YY525 pKa = 11.51ADD527 pKa = 3.31TYY529 pKa = 10.59LRR531 pKa = 11.84NVLDD535 pKa = 4.54GNPMSLDD542 pKa = 3.9SKK544 pKa = 10.75HH545 pKa = 6.22KK546 pKa = 10.68APMQITCPPLLITTNCNVLEE566 pKa = 4.17NNKK569 pKa = 8.69WKK571 pKa = 10.35YY572 pKa = 8.63LHH574 pKa = 6.59SRR576 pKa = 11.84LRR578 pKa = 11.84VYY580 pKa = 10.62NFHH583 pKa = 6.91NEE585 pKa = 3.89CPLNSRR591 pKa = 11.84GEE593 pKa = 4.14PEE595 pKa = 3.97FQLTKK600 pKa = 10.74VNWKK604 pKa = 10.57AFFNKK609 pKa = 9.44CWAKK613 pKa = 10.79LSLDD617 pKa = 4.02EE618 pKa = 5.34GDD620 pKa = 4.06EE621 pKa = 4.2GHH623 pKa = 6.59NGSPLQPLRR632 pKa = 11.84CVARR636 pKa = 11.84AADD639 pKa = 3.71EE640 pKa = 4.44ADD642 pKa = 3.13
MM1 pKa = 7.49DD2 pKa = 5.06TSPGTDD8 pKa = 3.51PLEE11 pKa = 4.7GGCNDD16 pKa = 3.24WVLLEE21 pKa = 4.11AAEE24 pKa = 4.54AGGSGIDD31 pKa = 3.57EE32 pKa = 4.39EE33 pKa = 4.44EE34 pKa = 5.66GEE36 pKa = 4.82DD37 pKa = 4.03EE38 pKa = 4.94DD39 pKa = 6.69DD40 pKa = 4.54SGEE43 pKa = 4.39DD44 pKa = 3.24MLDD47 pKa = 4.33FIDD50 pKa = 6.17DD51 pKa = 3.45SSTFQNAEE59 pKa = 3.78TQLYY63 pKa = 9.34CRR65 pKa = 11.84QLQVEE70 pKa = 4.24QQRR73 pKa = 11.84IDD75 pKa = 3.71DD76 pKa = 3.98EE77 pKa = 4.87RR78 pKa = 11.84VAQALKK84 pKa = 10.61RR85 pKa = 11.84KK86 pKa = 9.29FLDD89 pKa = 3.61SPKK92 pKa = 10.75SKK94 pKa = 10.57VDD96 pKa = 3.54SEE98 pKa = 4.24LSPRR102 pKa = 11.84LAAISLQEE110 pKa = 3.72QQRR113 pKa = 11.84RR114 pKa = 11.84GRR116 pKa = 11.84ARR118 pKa = 11.84RR119 pKa = 11.84RR120 pKa = 11.84LYY122 pKa = 10.58KK123 pKa = 10.64DD124 pKa = 2.99SGHH127 pKa = 6.54GDD129 pKa = 3.36SLEE132 pKa = 4.38AEE134 pKa = 4.57SEE136 pKa = 3.99EE137 pKa = 5.23SMVCRR142 pKa = 11.84NVQVPKK148 pKa = 10.11NQTMDD153 pKa = 3.21AVGAGEE159 pKa = 4.02GARR162 pKa = 11.84PKK164 pKa = 10.57EE165 pKa = 4.12VGNASTQEE173 pKa = 4.08VQCAGMSEE181 pKa = 4.42EE182 pKa = 4.53SACSVTQILRR192 pKa = 11.84AGKK195 pKa = 9.04PRR197 pKa = 11.84ALLLGVFKK205 pKa = 10.63EE206 pKa = 4.3AFGCSFADD214 pKa = 3.49LTRR217 pKa = 11.84CFKK220 pKa = 10.71SDD222 pKa = 3.09KK223 pKa = 9.94TVNDD227 pKa = 3.65DD228 pKa = 3.31WTCFIVGVPCSLEE241 pKa = 3.97GAISDD246 pKa = 4.06LLKK249 pKa = 10.58PSTLFTHH256 pKa = 5.85VTTATSKK263 pKa = 10.97YY264 pKa = 9.42GLTVLLLVQWKK275 pKa = 7.96TGKK278 pKa = 10.03SRR280 pKa = 11.84EE281 pKa = 4.38TVGNTLAGLLSVDD294 pKa = 3.48KK295 pKa = 10.75QQIVCEE301 pKa = 4.12PPKK304 pKa = 10.15IRR306 pKa = 11.84HH307 pKa = 6.0LGAAMFWYY315 pKa = 10.17KK316 pKa = 10.57KK317 pKa = 9.88AISSACVVTGEE328 pKa = 4.13MPVWILKK335 pKa = 9.21QVSIQDD341 pKa = 3.51QLGDD345 pKa = 3.43VCQFSLSVMIQWAYY359 pKa = 11.29DD360 pKa = 3.46NGHH363 pKa = 6.33DD364 pKa = 3.83TEE366 pKa = 4.17EE367 pKa = 5.15AIAYY371 pKa = 7.49EE372 pKa = 4.3YY373 pKa = 11.18AALADD378 pKa = 3.76EE379 pKa = 5.25DD380 pKa = 4.38KK381 pKa = 11.21NAEE384 pKa = 3.76AFLRR388 pKa = 11.84SNCQPKK394 pKa = 9.82YY395 pKa = 11.15VKK397 pKa = 10.49DD398 pKa = 3.74CANMLRR404 pKa = 11.84LYY406 pKa = 10.52KK407 pKa = 9.99RR408 pKa = 11.84AEE410 pKa = 3.82MRR412 pKa = 11.84KK413 pKa = 8.42MSMGQWIKK421 pKa = 10.58HH422 pKa = 5.65KK423 pKa = 10.28CDD425 pKa = 3.22AVQGEE430 pKa = 4.77GDD432 pKa = 3.62WKK434 pKa = 10.9HH435 pKa = 5.91VMNFLKK441 pKa = 10.72FQGIEE446 pKa = 3.78IMPFLIDD453 pKa = 4.22FRR455 pKa = 11.84LFLKK459 pKa = 10.08GIPKK463 pKa = 9.5RR464 pKa = 11.84NCMVLYY470 pKa = 10.16GPPNTGKK477 pKa = 10.5SLFAMSLIHH486 pKa = 7.02LLGGKK491 pKa = 8.88VLSYY495 pKa = 11.26VNSSSHH501 pKa = 6.44FWLQPVADD509 pKa = 4.63CKK511 pKa = 11.08VALIDD516 pKa = 5.26DD517 pKa = 4.58ATASTWDD524 pKa = 3.48YY525 pKa = 11.51ADD527 pKa = 3.31TYY529 pKa = 10.59LRR531 pKa = 11.84NVLDD535 pKa = 4.54GNPMSLDD542 pKa = 3.9SKK544 pKa = 10.75HH545 pKa = 6.22KK546 pKa = 10.68APMQITCPPLLITTNCNVLEE566 pKa = 4.17NNKK569 pKa = 8.69WKK571 pKa = 10.35YY572 pKa = 8.63LHH574 pKa = 6.59SRR576 pKa = 11.84LRR578 pKa = 11.84VYY580 pKa = 10.62NFHH583 pKa = 6.91NEE585 pKa = 3.89CPLNSRR591 pKa = 11.84GEE593 pKa = 4.14PEE595 pKa = 3.97FQLTKK600 pKa = 10.74VNWKK604 pKa = 10.57AFFNKK609 pKa = 9.44CWAKK613 pKa = 10.79LSLDD617 pKa = 4.02EE618 pKa = 5.34GDD620 pKa = 4.06EE621 pKa = 4.2GHH623 pKa = 6.59NGSPLQPLRR632 pKa = 11.84CVARR636 pKa = 11.84AADD639 pKa = 3.71EE640 pKa = 4.44ADD642 pKa = 3.13
Molecular weight: 71.64 kDa
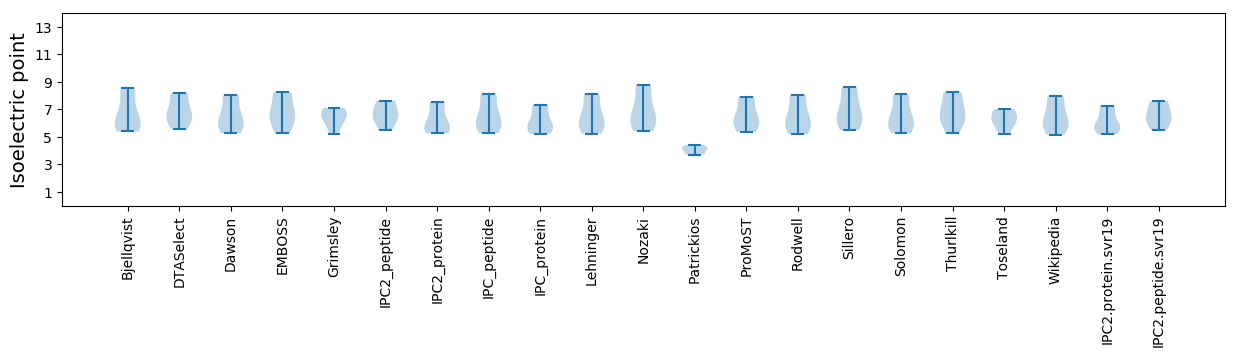
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F2VIS7|F2VIS7_9PAPI E4 protein (Fragment) OS=Lagenorhynchus acutus papillomavirus OX=706530 GN=E4 PE=4 SV=1
MM1 pKa = 7.7KK2 pKa = 7.83EE3 pKa = 4.08TKK5 pKa = 9.57DD6 pKa = 3.47TMEE9 pKa = 4.24VLCSRR14 pKa = 11.84LDD16 pKa = 3.6ALQEE20 pKa = 3.93QQMKK24 pKa = 10.69LIEE27 pKa = 4.43TDD29 pKa = 4.55DD30 pKa = 4.08GDD32 pKa = 4.23LEE34 pKa = 4.57NVLLYY39 pKa = 10.63FGLLRR44 pKa = 11.84KK45 pKa = 8.29EE46 pKa = 4.27TMLLYY51 pKa = 10.38AANEE55 pKa = 4.08RR56 pKa = 11.84GHH58 pKa = 6.39KK59 pKa = 10.06KK60 pKa = 10.45VGFTMVPNKK69 pKa = 9.76KK70 pKa = 10.08ACEE73 pKa = 3.86EE74 pKa = 3.94NAKK77 pKa = 10.3KK78 pKa = 10.23CIRR81 pKa = 11.84MHH83 pKa = 6.51LAVSSLLSSLFAKK96 pKa = 9.26EE97 pKa = 4.1KK98 pKa = 9.92WFLSQVSYY106 pKa = 11.49EE107 pKa = 4.28MFQTPPVEE115 pKa = 4.28TFKK118 pKa = 11.36KK119 pKa = 9.52EE120 pKa = 3.69GRR122 pKa = 11.84TVTVTFDD129 pKa = 4.45DD130 pKa = 4.83DD131 pKa = 3.92EE132 pKa = 4.8QNSMEE137 pKa = 4.38YY138 pKa = 10.56VCWTRR143 pKa = 11.84IYY145 pKa = 10.37CQLPNGEE152 pKa = 4.27WGCYY156 pKa = 8.43CAVVCYY162 pKa = 10.23EE163 pKa = 4.27GIYY166 pKa = 10.42YY167 pKa = 10.35DD168 pKa = 3.69VEE170 pKa = 4.15GEE172 pKa = 3.86KK173 pKa = 10.45RR174 pKa = 11.84FYY176 pKa = 11.51VDD178 pKa = 5.11FLSEE182 pKa = 3.88ALKK185 pKa = 11.11YY186 pKa = 9.33GTGEE190 pKa = 3.83KK191 pKa = 8.11WTVLYY196 pKa = 10.43DD197 pKa = 3.72GKK199 pKa = 10.9AITDD203 pKa = 3.8CTLVSSTSTTSPTSALGYY221 pKa = 9.08PILSSTPGNPGGEE234 pKa = 4.05PAHH237 pKa = 5.67TKK239 pKa = 9.94RR240 pKa = 11.84RR241 pKa = 11.84RR242 pKa = 11.84KK243 pKa = 8.56PQTRR247 pKa = 11.84RR248 pKa = 11.84LLGPPPSPPPSPPPGRR264 pKa = 11.84PPPPRR269 pKa = 11.84PPTPPPRR276 pKa = 11.84PPATPPSPLATPGSAKK292 pKa = 10.02RR293 pKa = 11.84HH294 pKa = 5.92RR295 pKa = 11.84EE296 pKa = 3.46TSAAAGAEE304 pKa = 4.1GRR306 pKa = 11.84GGSGDD311 pKa = 3.51SFGGVSEE318 pKa = 4.62CDD320 pKa = 3.54CNRR323 pKa = 11.84DD324 pKa = 3.05SGKK327 pKa = 9.99RR328 pKa = 11.84RR329 pKa = 11.84RR330 pKa = 11.84TDD332 pKa = 3.3PLIPCLLIGGGANQVKK348 pKa = 10.01CLRR351 pKa = 11.84HH352 pKa = 5.31RR353 pKa = 11.84LKK355 pKa = 10.94VQFFGLYY362 pKa = 9.91SNCSTTWSWVGHH374 pKa = 5.92SGSTPNHH381 pKa = 6.64RR382 pKa = 11.84ICVSFLNEE390 pKa = 3.55EE391 pKa = 3.86QRR393 pKa = 11.84NIFLNFVPLPGTVTVAPAEE412 pKa = 4.24LPFF415 pKa = 5.72
MM1 pKa = 7.7KK2 pKa = 7.83EE3 pKa = 4.08TKK5 pKa = 9.57DD6 pKa = 3.47TMEE9 pKa = 4.24VLCSRR14 pKa = 11.84LDD16 pKa = 3.6ALQEE20 pKa = 3.93QQMKK24 pKa = 10.69LIEE27 pKa = 4.43TDD29 pKa = 4.55DD30 pKa = 4.08GDD32 pKa = 4.23LEE34 pKa = 4.57NVLLYY39 pKa = 10.63FGLLRR44 pKa = 11.84KK45 pKa = 8.29EE46 pKa = 4.27TMLLYY51 pKa = 10.38AANEE55 pKa = 4.08RR56 pKa = 11.84GHH58 pKa = 6.39KK59 pKa = 10.06KK60 pKa = 10.45VGFTMVPNKK69 pKa = 9.76KK70 pKa = 10.08ACEE73 pKa = 3.86EE74 pKa = 3.94NAKK77 pKa = 10.3KK78 pKa = 10.23CIRR81 pKa = 11.84MHH83 pKa = 6.51LAVSSLLSSLFAKK96 pKa = 9.26EE97 pKa = 4.1KK98 pKa = 9.92WFLSQVSYY106 pKa = 11.49EE107 pKa = 4.28MFQTPPVEE115 pKa = 4.28TFKK118 pKa = 11.36KK119 pKa = 9.52EE120 pKa = 3.69GRR122 pKa = 11.84TVTVTFDD129 pKa = 4.45DD130 pKa = 4.83DD131 pKa = 3.92EE132 pKa = 4.8QNSMEE137 pKa = 4.38YY138 pKa = 10.56VCWTRR143 pKa = 11.84IYY145 pKa = 10.37CQLPNGEE152 pKa = 4.27WGCYY156 pKa = 8.43CAVVCYY162 pKa = 10.23EE163 pKa = 4.27GIYY166 pKa = 10.42YY167 pKa = 10.35DD168 pKa = 3.69VEE170 pKa = 4.15GEE172 pKa = 3.86KK173 pKa = 10.45RR174 pKa = 11.84FYY176 pKa = 11.51VDD178 pKa = 5.11FLSEE182 pKa = 3.88ALKK185 pKa = 11.11YY186 pKa = 9.33GTGEE190 pKa = 3.83KK191 pKa = 8.11WTVLYY196 pKa = 10.43DD197 pKa = 3.72GKK199 pKa = 10.9AITDD203 pKa = 3.8CTLVSSTSTTSPTSALGYY221 pKa = 9.08PILSSTPGNPGGEE234 pKa = 4.05PAHH237 pKa = 5.67TKK239 pKa = 9.94RR240 pKa = 11.84RR241 pKa = 11.84RR242 pKa = 11.84KK243 pKa = 8.56PQTRR247 pKa = 11.84RR248 pKa = 11.84LLGPPPSPPPSPPPGRR264 pKa = 11.84PPPPRR269 pKa = 11.84PPTPPPRR276 pKa = 11.84PPATPPSPLATPGSAKK292 pKa = 10.02RR293 pKa = 11.84HH294 pKa = 5.92RR295 pKa = 11.84EE296 pKa = 3.46TSAAAGAEE304 pKa = 4.1GRR306 pKa = 11.84GGSGDD311 pKa = 3.51SFGGVSEE318 pKa = 4.62CDD320 pKa = 3.54CNRR323 pKa = 11.84DD324 pKa = 3.05SGKK327 pKa = 9.99RR328 pKa = 11.84RR329 pKa = 11.84RR330 pKa = 11.84TDD332 pKa = 3.3PLIPCLLIGGGANQVKK348 pKa = 10.01CLRR351 pKa = 11.84HH352 pKa = 5.31RR353 pKa = 11.84LKK355 pKa = 10.94VQFFGLYY362 pKa = 9.91SNCSTTWSWVGHH374 pKa = 5.92SGSTPNHH381 pKa = 6.64RR382 pKa = 11.84ICVSFLNEE390 pKa = 3.55EE391 pKa = 3.86QRR393 pKa = 11.84NIFLNFVPLPGTVTVAPAEE412 pKa = 4.24LPFF415 pKa = 5.72
Molecular weight: 45.94 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2460 |
137 |
642 |
410.0 |
45.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.626 ± 0.513 | 2.48 ± 0.695 |
5.935 ± 0.535 | 5.813 ± 0.626 |
3.78 ± 0.334 | 6.951 ± 0.384 |
2.846 ± 1.027 | 3.699 ± 0.452 |
4.512 ± 0.921 | 9.431 ± 0.449 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.789 ± 0.41 | 3.821 ± 0.502 |
7.073 ± 1.058 | 3.577 ± 0.675 |
6.179 ± 0.536 | 8.618 ± 0.914 |
5.894 ± 0.573 | 6.504 ± 0.377 |
1.504 ± 0.229 | 2.967 ± 0.224 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
