
Human papillomavirus type 144
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus; Gammapapillomavirus 17
Average proteome isoelectric point is 6.13
Get precalculated fractions of proteins
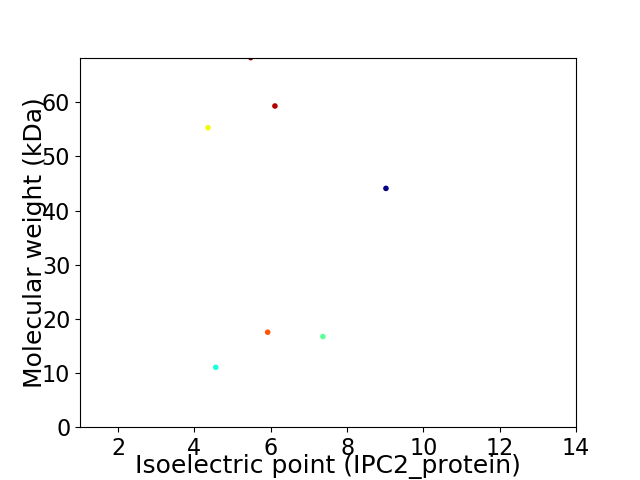
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I3P6Q1|I3P6Q1_9PAPI Major capsid protein L1 OS=Human papillomavirus type 144 OX=1070417 GN=L1 PE=3 SV=1
MM1 pKa = 7.7DD2 pKa = 4.49AVYY5 pKa = 9.81PRR7 pKa = 11.84RR8 pKa = 11.84IKK10 pKa = 10.46RR11 pKa = 11.84DD12 pKa = 3.52SVDD15 pKa = 3.65NLYY18 pKa = 10.02KK19 pKa = 10.42QCAITGNCPPDD30 pKa = 3.48VKK32 pKa = 11.12DD33 pKa = 3.47KK34 pKa = 11.92VEE36 pKa = 4.25GNTLADD42 pKa = 3.57KK43 pKa = 10.53LLKK46 pKa = 10.14IFSSILFFGGLGIGTGKK63 pKa = 8.8GTGGTLGYY71 pKa = 10.44RR72 pKa = 11.84PLTPSTPRR80 pKa = 11.84VLPEE84 pKa = 3.72GTIVRR89 pKa = 11.84PTPPIDD95 pKa = 4.46PIGPPDD101 pKa = 4.77IISIDD106 pKa = 4.16AIDD109 pKa = 4.15PAGSSIVPLQEE120 pKa = 4.86AGGLPDD126 pKa = 3.34SGVIEE131 pKa = 4.82EE132 pKa = 4.61IPLVEE137 pKa = 4.81LGQPSVVSTSDD148 pKa = 3.99PISDD152 pKa = 3.07ISLPEE157 pKa = 3.84STPTVVTTADD167 pKa = 3.51DD168 pKa = 3.58TAVAVLDD175 pKa = 4.07ISPAQPAPKK184 pKa = 9.5RR185 pKa = 11.84IVVEE189 pKa = 4.5GRR191 pKa = 11.84VKK193 pKa = 10.35PYY195 pKa = 10.53QLVHH199 pKa = 6.43ISSLAATAPEE209 pKa = 4.2GADD212 pKa = 2.69INVFVDD218 pKa = 2.77SHH220 pKa = 7.47FDD222 pKa = 3.28GDD224 pKa = 4.38VIGGQYY230 pKa = 10.64EE231 pKa = 4.55EE232 pKa = 5.36IPLEE236 pKa = 5.04DD237 pKa = 3.72ISSINPPKK245 pKa = 10.36TSTPQGTVPRR255 pKa = 11.84LTQRR259 pKa = 11.84ARR261 pKa = 11.84NFYY264 pKa = 9.67GRR266 pKa = 11.84RR267 pKa = 11.84VQQVPTTNVDD277 pKa = 3.92FLSRR281 pKa = 11.84PSRR284 pKa = 11.84LVQFEE289 pKa = 4.27NPAFDD294 pKa = 5.23ADD296 pKa = 5.14DD297 pKa = 3.26ITLQFEE303 pKa = 3.99QDD305 pKa = 3.65LEE307 pKa = 4.38QVLAAPDD314 pKa = 3.64TDD316 pKa = 3.84FQDD319 pKa = 3.08IRR321 pKa = 11.84KK322 pKa = 8.88LSSVSYY328 pKa = 10.71SDD330 pKa = 3.05VQGRR334 pKa = 11.84VRR336 pKa = 11.84VSRR339 pKa = 11.84LGTRR343 pKa = 11.84GTIQTRR349 pKa = 11.84SGVLIGEE356 pKa = 4.33DD357 pKa = 3.14VHH359 pKa = 8.34FYY361 pKa = 11.17FDD363 pKa = 5.52LSTIDD368 pKa = 3.27NSGAIEE374 pKa = 3.94LQPLGEE380 pKa = 4.22HH381 pKa = 6.2TGDD384 pKa = 3.25VTLVDD389 pKa = 4.53GLAEE393 pKa = 4.13SSFIDD398 pKa = 3.86AVDD401 pKa = 3.62NNEE404 pKa = 4.54LDD406 pKa = 5.01LEE408 pKa = 4.36TLLLDD413 pKa = 3.92EE414 pKa = 4.25QTEE417 pKa = 4.36DD418 pKa = 3.74FSNSHH423 pKa = 7.11LIITGTSRR431 pKa = 11.84QNTLSIPTIPPSVSLKK447 pKa = 10.61IFVDD451 pKa = 3.92DD452 pKa = 4.02VGDD455 pKa = 3.87NLFVSYY461 pKa = 9.78PKK463 pKa = 10.46SHH465 pKa = 6.56TPSDD469 pKa = 3.81TVFVDD474 pKa = 4.16TPFTPLDD481 pKa = 3.5PAILIDD487 pKa = 3.81VASDD491 pKa = 3.16TYY493 pKa = 11.06YY494 pKa = 10.31IHH496 pKa = 8.01PSLLKK501 pKa = 10.13KK502 pKa = 10.22GKK504 pKa = 8.97KK505 pKa = 9.3RR506 pKa = 11.84KK507 pKa = 9.96YY508 pKa = 10.52SDD510 pKa = 2.79IFF512 pKa = 4.01
MM1 pKa = 7.7DD2 pKa = 4.49AVYY5 pKa = 9.81PRR7 pKa = 11.84RR8 pKa = 11.84IKK10 pKa = 10.46RR11 pKa = 11.84DD12 pKa = 3.52SVDD15 pKa = 3.65NLYY18 pKa = 10.02KK19 pKa = 10.42QCAITGNCPPDD30 pKa = 3.48VKK32 pKa = 11.12DD33 pKa = 3.47KK34 pKa = 11.92VEE36 pKa = 4.25GNTLADD42 pKa = 3.57KK43 pKa = 10.53LLKK46 pKa = 10.14IFSSILFFGGLGIGTGKK63 pKa = 8.8GTGGTLGYY71 pKa = 10.44RR72 pKa = 11.84PLTPSTPRR80 pKa = 11.84VLPEE84 pKa = 3.72GTIVRR89 pKa = 11.84PTPPIDD95 pKa = 4.46PIGPPDD101 pKa = 4.77IISIDD106 pKa = 4.16AIDD109 pKa = 4.15PAGSSIVPLQEE120 pKa = 4.86AGGLPDD126 pKa = 3.34SGVIEE131 pKa = 4.82EE132 pKa = 4.61IPLVEE137 pKa = 4.81LGQPSVVSTSDD148 pKa = 3.99PISDD152 pKa = 3.07ISLPEE157 pKa = 3.84STPTVVTTADD167 pKa = 3.51DD168 pKa = 3.58TAVAVLDD175 pKa = 4.07ISPAQPAPKK184 pKa = 9.5RR185 pKa = 11.84IVVEE189 pKa = 4.5GRR191 pKa = 11.84VKK193 pKa = 10.35PYY195 pKa = 10.53QLVHH199 pKa = 6.43ISSLAATAPEE209 pKa = 4.2GADD212 pKa = 2.69INVFVDD218 pKa = 2.77SHH220 pKa = 7.47FDD222 pKa = 3.28GDD224 pKa = 4.38VIGGQYY230 pKa = 10.64EE231 pKa = 4.55EE232 pKa = 5.36IPLEE236 pKa = 5.04DD237 pKa = 3.72ISSINPPKK245 pKa = 10.36TSTPQGTVPRR255 pKa = 11.84LTQRR259 pKa = 11.84ARR261 pKa = 11.84NFYY264 pKa = 9.67GRR266 pKa = 11.84RR267 pKa = 11.84VQQVPTTNVDD277 pKa = 3.92FLSRR281 pKa = 11.84PSRR284 pKa = 11.84LVQFEE289 pKa = 4.27NPAFDD294 pKa = 5.23ADD296 pKa = 5.14DD297 pKa = 3.26ITLQFEE303 pKa = 3.99QDD305 pKa = 3.65LEE307 pKa = 4.38QVLAAPDD314 pKa = 3.64TDD316 pKa = 3.84FQDD319 pKa = 3.08IRR321 pKa = 11.84KK322 pKa = 8.88LSSVSYY328 pKa = 10.71SDD330 pKa = 3.05VQGRR334 pKa = 11.84VRR336 pKa = 11.84VSRR339 pKa = 11.84LGTRR343 pKa = 11.84GTIQTRR349 pKa = 11.84SGVLIGEE356 pKa = 4.33DD357 pKa = 3.14VHH359 pKa = 8.34FYY361 pKa = 11.17FDD363 pKa = 5.52LSTIDD368 pKa = 3.27NSGAIEE374 pKa = 3.94LQPLGEE380 pKa = 4.22HH381 pKa = 6.2TGDD384 pKa = 3.25VTLVDD389 pKa = 4.53GLAEE393 pKa = 4.13SSFIDD398 pKa = 3.86AVDD401 pKa = 3.62NNEE404 pKa = 4.54LDD406 pKa = 5.01LEE408 pKa = 4.36TLLLDD413 pKa = 3.92EE414 pKa = 4.25QTEE417 pKa = 4.36DD418 pKa = 3.74FSNSHH423 pKa = 7.11LIITGTSRR431 pKa = 11.84QNTLSIPTIPPSVSLKK447 pKa = 10.61IFVDD451 pKa = 3.92DD452 pKa = 4.02VGDD455 pKa = 3.87NLFVSYY461 pKa = 9.78PKK463 pKa = 10.46SHH465 pKa = 6.56TPSDD469 pKa = 3.81TVFVDD474 pKa = 4.16TPFTPLDD481 pKa = 3.5PAILIDD487 pKa = 3.81VASDD491 pKa = 3.16TYY493 pKa = 11.06YY494 pKa = 10.31IHH496 pKa = 8.01PSLLKK501 pKa = 10.13KK502 pKa = 10.22GKK504 pKa = 8.97KK505 pKa = 9.3RR506 pKa = 11.84KK507 pKa = 9.96YY508 pKa = 10.52SDD510 pKa = 2.79IFF512 pKa = 4.01
Molecular weight: 55.3 kDa
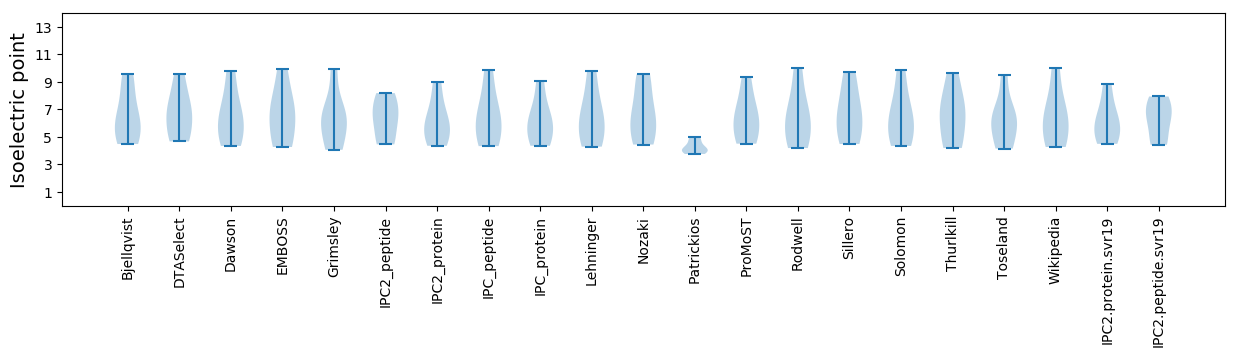
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I3P6P9|I3P6P9_9PAPI Uncharacterized protein OS=Human papillomavirus type 144 OX=1070417 GN=E4 PE=4 SV=1
MM1 pKa = 7.28ATLLRR6 pKa = 11.84DD7 pKa = 3.75RR8 pKa = 11.84FRR10 pKa = 11.84ALQEE14 pKa = 3.97ALMEE18 pKa = 4.6SYY20 pKa = 10.95EE21 pKa = 4.28SGATDD26 pKa = 3.47LHH28 pKa = 6.16SQLTYY33 pKa = 7.96WHH35 pKa = 7.16LVRR38 pKa = 11.84KK39 pKa = 8.09EE40 pKa = 3.61QAVLYY45 pKa = 10.05KK46 pKa = 10.51ARR48 pKa = 11.84QDD50 pKa = 3.62GLRR53 pKa = 11.84TLGLQPVPTLQVSEE67 pKa = 4.33YY68 pKa = 8.98RR69 pKa = 11.84AKK71 pKa = 10.55EE72 pKa = 4.27AISMTLLLRR81 pKa = 11.84TFIKK85 pKa = 10.61SQFARR90 pKa = 11.84EE91 pKa = 3.94SWTLQDD97 pKa = 3.59VSAEE101 pKa = 4.24LVLHH105 pKa = 6.47TKK107 pKa = 9.87PKK109 pKa = 9.1NTFKK113 pKa = 10.94KK114 pKa = 9.58RR115 pKa = 11.84GYY117 pKa = 7.4TVEE120 pKa = 3.99VLFDD124 pKa = 4.48HH125 pKa = 7.58DD126 pKa = 3.95PMKK129 pKa = 10.92AFPYY133 pKa = 10.22TNFTEE138 pKa = 5.46IYY140 pKa = 8.41YY141 pKa = 9.91QDD143 pKa = 4.24EE144 pKa = 4.13NEE146 pKa = 4.23MWHH149 pKa = 5.43KK150 pKa = 9.93TEE152 pKa = 4.99GKK154 pKa = 9.59VDD156 pKa = 3.8YY157 pKa = 11.09DD158 pKa = 3.27GMYY161 pKa = 10.56YY162 pKa = 10.25EE163 pKa = 5.13EE164 pKa = 4.89PNGDD168 pKa = 2.77RR169 pKa = 11.84VYY171 pKa = 11.03FQLFDD176 pKa = 4.4EE177 pKa = 5.1DD178 pKa = 3.82AQTYY182 pKa = 7.87GQSGQWTVNYY192 pKa = 10.35KK193 pKa = 8.71NTTLSPPVTSSTRR206 pKa = 11.84PTSSASTTGVGGTTDD221 pKa = 3.09TEE223 pKa = 4.35AQQKK227 pKa = 9.04VPSGGLQTAEE237 pKa = 4.28VASTSSPSRR246 pKa = 11.84RR247 pKa = 11.84RR248 pKa = 11.84RR249 pKa = 11.84RR250 pKa = 11.84EE251 pKa = 3.56QQRR254 pKa = 11.84EE255 pKa = 4.1SPVPSKK261 pKa = 10.45RR262 pKa = 11.84RR263 pKa = 11.84RR264 pKa = 11.84RR265 pKa = 11.84SGSTGQGTVPTPGEE279 pKa = 3.69VGGRR283 pKa = 11.84HH284 pKa = 4.19RR285 pKa = 11.84TVARR289 pKa = 11.84TGLSRR294 pKa = 11.84IRR296 pKa = 11.84RR297 pKa = 11.84LQEE300 pKa = 3.4EE301 pKa = 4.25ARR303 pKa = 11.84DD304 pKa = 3.74PPIAIIKK311 pKa = 10.28GPANPLKK318 pKa = 10.43CWRR321 pKa = 11.84NRR323 pKa = 11.84CNQKK327 pKa = 9.66YY328 pKa = 10.35ASFFVSMSTAWNWVNDD344 pKa = 3.84DD345 pKa = 3.34STKK348 pKa = 10.15QEE350 pKa = 3.75RR351 pKa = 11.84SRR353 pKa = 11.84LLVAFASTAQRR364 pKa = 11.84DD365 pKa = 4.0MFVKK369 pKa = 10.5LVNFPRR375 pKa = 11.84GTTYY379 pKa = 11.46AFGHH383 pKa = 6.93LDD385 pKa = 3.39SLL387 pKa = 4.57
MM1 pKa = 7.28ATLLRR6 pKa = 11.84DD7 pKa = 3.75RR8 pKa = 11.84FRR10 pKa = 11.84ALQEE14 pKa = 3.97ALMEE18 pKa = 4.6SYY20 pKa = 10.95EE21 pKa = 4.28SGATDD26 pKa = 3.47LHH28 pKa = 6.16SQLTYY33 pKa = 7.96WHH35 pKa = 7.16LVRR38 pKa = 11.84KK39 pKa = 8.09EE40 pKa = 3.61QAVLYY45 pKa = 10.05KK46 pKa = 10.51ARR48 pKa = 11.84QDD50 pKa = 3.62GLRR53 pKa = 11.84TLGLQPVPTLQVSEE67 pKa = 4.33YY68 pKa = 8.98RR69 pKa = 11.84AKK71 pKa = 10.55EE72 pKa = 4.27AISMTLLLRR81 pKa = 11.84TFIKK85 pKa = 10.61SQFARR90 pKa = 11.84EE91 pKa = 3.94SWTLQDD97 pKa = 3.59VSAEE101 pKa = 4.24LVLHH105 pKa = 6.47TKK107 pKa = 9.87PKK109 pKa = 9.1NTFKK113 pKa = 10.94KK114 pKa = 9.58RR115 pKa = 11.84GYY117 pKa = 7.4TVEE120 pKa = 3.99VLFDD124 pKa = 4.48HH125 pKa = 7.58DD126 pKa = 3.95PMKK129 pKa = 10.92AFPYY133 pKa = 10.22TNFTEE138 pKa = 5.46IYY140 pKa = 8.41YY141 pKa = 9.91QDD143 pKa = 4.24EE144 pKa = 4.13NEE146 pKa = 4.23MWHH149 pKa = 5.43KK150 pKa = 9.93TEE152 pKa = 4.99GKK154 pKa = 9.59VDD156 pKa = 3.8YY157 pKa = 11.09DD158 pKa = 3.27GMYY161 pKa = 10.56YY162 pKa = 10.25EE163 pKa = 5.13EE164 pKa = 4.89PNGDD168 pKa = 2.77RR169 pKa = 11.84VYY171 pKa = 11.03FQLFDD176 pKa = 4.4EE177 pKa = 5.1DD178 pKa = 3.82AQTYY182 pKa = 7.87GQSGQWTVNYY192 pKa = 10.35KK193 pKa = 8.71NTTLSPPVTSSTRR206 pKa = 11.84PTSSASTTGVGGTTDD221 pKa = 3.09TEE223 pKa = 4.35AQQKK227 pKa = 9.04VPSGGLQTAEE237 pKa = 4.28VASTSSPSRR246 pKa = 11.84RR247 pKa = 11.84RR248 pKa = 11.84RR249 pKa = 11.84RR250 pKa = 11.84EE251 pKa = 3.56QQRR254 pKa = 11.84EE255 pKa = 4.1SPVPSKK261 pKa = 10.45RR262 pKa = 11.84RR263 pKa = 11.84RR264 pKa = 11.84RR265 pKa = 11.84SGSTGQGTVPTPGEE279 pKa = 3.69VGGRR283 pKa = 11.84HH284 pKa = 4.19RR285 pKa = 11.84TVARR289 pKa = 11.84TGLSRR294 pKa = 11.84IRR296 pKa = 11.84RR297 pKa = 11.84LQEE300 pKa = 3.4EE301 pKa = 4.25ARR303 pKa = 11.84DD304 pKa = 3.74PPIAIIKK311 pKa = 10.28GPANPLKK318 pKa = 10.43CWRR321 pKa = 11.84NRR323 pKa = 11.84CNQKK327 pKa = 9.66YY328 pKa = 10.35ASFFVSMSTAWNWVNDD344 pKa = 3.84DD345 pKa = 3.34STKK348 pKa = 10.15QEE350 pKa = 3.75RR351 pKa = 11.84SRR353 pKa = 11.84LLVAFASTAQRR364 pKa = 11.84DD365 pKa = 4.0MFVKK369 pKa = 10.5LVNFPRR375 pKa = 11.84GTTYY379 pKa = 11.46AFGHH383 pKa = 6.93LDD385 pKa = 3.39SLL387 pKa = 4.57
Molecular weight: 44.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2404 |
99 |
599 |
343.4 |
38.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.699 ± 0.314 | 1.913 ± 0.709 |
6.78 ± 0.686 | 6.24 ± 0.675 |
4.908 ± 0.475 | 5.366 ± 0.631 |
2.121 ± 0.238 | 5.491 ± 0.771 |
5.116 ± 0.576 | 9.235 ± 0.776 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.83 ± 0.418 | 4.576 ± 0.909 |
6.115 ± 0.867 | 4.908 ± 0.376 |
6.198 ± 0.921 | 7.404 ± 0.725 |
6.073 ± 1.149 | 5.99 ± 0.7 |
1.206 ± 0.316 | 2.829 ± 0.437 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
