
Intestinimonas butyriciproducens
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Eubacteriales incertae sedis; Intestinimonas
Average proteome isoelectric point is 6.27
Get precalculated fractions of proteins
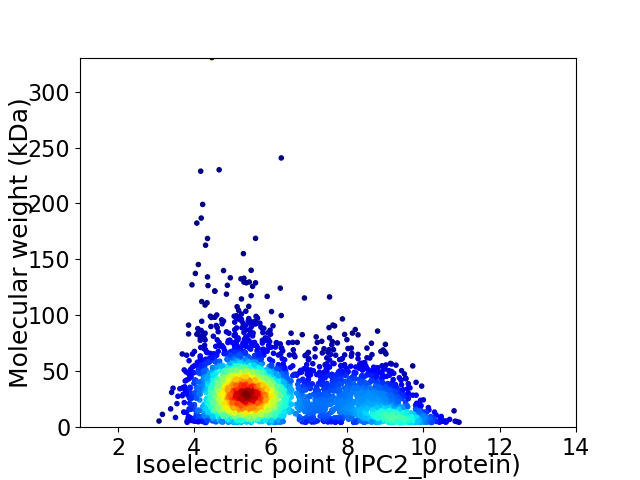
Virtual 2D-PAGE plot for 3358 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
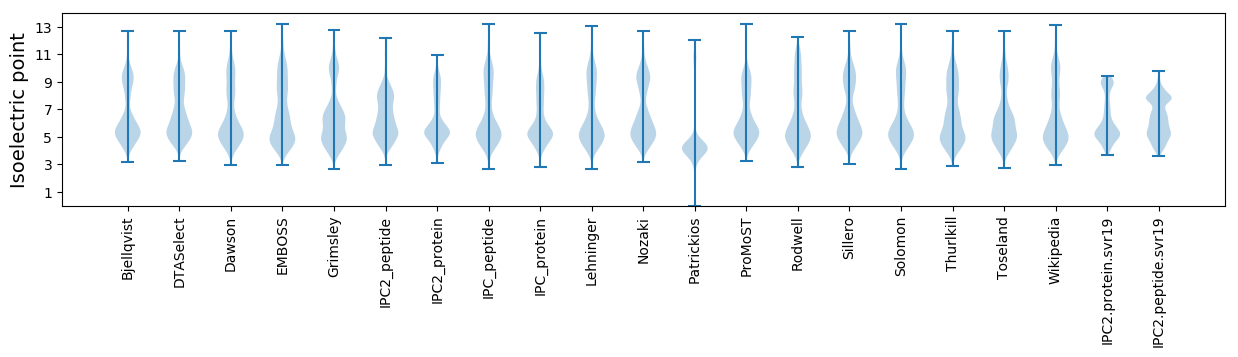
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0S2W2M6|A0A0S2W2M6_9FIRM Uncharacterized protein OS=Intestinimonas butyriciproducens OX=1297617 GN=IB211_01126c PE=4 SV=1
MM1 pKa = 7.42KK2 pKa = 10.67NKK4 pKa = 9.59IKK6 pKa = 10.68KK7 pKa = 9.93SFAAILALAMLLSLAACGGKK27 pKa = 10.14DD28 pKa = 3.62KK29 pKa = 11.42GSDD32 pKa = 3.38GDD34 pKa = 4.46GSDD37 pKa = 3.29KK38 pKa = 11.29TEE40 pKa = 4.16TIVLGKK46 pKa = 8.19YY47 pKa = 6.62TAVFKK52 pKa = 10.87GYY54 pKa = 8.09TLTKK58 pKa = 10.45DD59 pKa = 4.32DD60 pKa = 4.69EE61 pKa = 5.2GNDD64 pKa = 3.69AMVLTYY70 pKa = 10.51DD71 pKa = 3.64YY72 pKa = 11.25TNGSKK77 pKa = 10.69DD78 pKa = 3.55EE79 pKa = 4.25QSFAWAFLFEE89 pKa = 4.12ATQAGEE95 pKa = 4.14EE96 pKa = 4.62LGSPMYY102 pKa = 10.54DD103 pKa = 3.11EE104 pKa = 5.57NNNQITKK111 pKa = 10.44NYY113 pKa = 8.24EE114 pKa = 3.83EE115 pKa = 4.6KK116 pKa = 10.68VKK118 pKa = 10.53KK119 pKa = 10.52GQTLEE124 pKa = 3.89VAIALTLVNTTDD136 pKa = 3.32DD137 pKa = 4.15VIIVFSDD144 pKa = 3.77FDD146 pKa = 3.73DD147 pKa = 4.43HH148 pKa = 7.67EE149 pKa = 4.4YY150 pKa = 9.09TQTVKK155 pKa = 10.96LSGSGVQDD163 pKa = 3.34SGPEE167 pKa = 3.87EE168 pKa = 4.03AGSYY172 pKa = 8.63QLYY175 pKa = 10.6EE176 pKa = 3.9MTFEE180 pKa = 5.11GDD182 pKa = 3.69VFDD185 pKa = 5.78HH186 pKa = 7.46DD187 pKa = 4.23FLEE190 pKa = 4.65TSGFEE195 pKa = 4.36LYY197 pKa = 10.56TMEE200 pKa = 5.03LAADD204 pKa = 3.82GTGWLQIDD212 pKa = 3.98GRR214 pKa = 11.84GSLTWGDD221 pKa = 3.63GKK223 pKa = 9.47ITIDD227 pKa = 4.14GSDD230 pKa = 3.11AVYY233 pKa = 9.44TYY235 pKa = 9.56TYY237 pKa = 9.28KK238 pKa = 11.07DD239 pKa = 3.22GMIVLEE245 pKa = 4.25EE246 pKa = 4.21STGEE250 pKa = 3.98SMTFQKK256 pKa = 9.77TGAAGTSGGLKK267 pKa = 10.07PVGGDD272 pKa = 3.29GSDD275 pKa = 3.92ANLTAYY281 pKa = 8.18QQYY284 pKa = 10.37WNGDD288 pKa = 3.06WYY290 pKa = 11.17GWTAYY295 pKa = 11.21SNGTNDD301 pKa = 3.27YY302 pKa = 10.86SSLDD306 pKa = 3.86GFYY309 pKa = 8.99MDD311 pKa = 4.01CMARR315 pKa = 11.84IEE317 pKa = 4.01IDD319 pKa = 3.6EE320 pKa = 4.49NGEE323 pKa = 3.79GALILWDD330 pKa = 3.75EE331 pKa = 4.5ATSVDD336 pKa = 4.11DD337 pKa = 3.85PRR339 pKa = 11.84AGVTVCISDD348 pKa = 3.44GGFEE352 pKa = 3.95QGRR355 pKa = 11.84LKK357 pKa = 11.05SEE359 pKa = 4.83DD360 pKa = 2.96GWFSDD365 pKa = 4.16GDD367 pKa = 3.73VGNADD372 pKa = 3.62WLVDD376 pKa = 3.89PDD378 pKa = 3.79GLGYY382 pKa = 10.93EE383 pKa = 4.06NLICIEE389 pKa = 4.34GSYY392 pKa = 10.66KK393 pKa = 11.08DD394 pKa = 3.82PDD396 pKa = 4.19DD397 pKa = 4.34FFSGYY402 pKa = 10.79DD403 pKa = 3.22YY404 pKa = 10.96TIYY407 pKa = 10.54LRR409 pKa = 11.84PWGLDD414 pKa = 2.91WADD417 pKa = 3.82IEE419 pKa = 5.27ADD421 pKa = 3.83QPDD424 pKa = 4.96GLPDD428 pKa = 3.7HH429 pKa = 6.83YY430 pKa = 11.22DD431 pKa = 3.04WYY433 pKa = 11.25LSAIQTGAKK442 pKa = 8.52MPDD445 pKa = 3.92VIGGDD450 pKa = 3.84YY451 pKa = 11.07GGLAPGEE458 pKa = 4.41VPDD461 pKa = 5.16SQMEE465 pKa = 4.17PQGGNDD471 pKa = 3.7PNASAVDD478 pKa = 4.18GTWQAPVYY486 pKa = 10.67GDD488 pKa = 3.62YY489 pKa = 11.36GLSKK493 pKa = 10.91ASATGVVPMTRR504 pKa = 11.84DD505 pKa = 3.38EE506 pKa = 4.18LVDD509 pKa = 4.54RR510 pKa = 11.84IGFYY514 pKa = 10.97ADD516 pKa = 3.5SQIGSAPYY524 pKa = 10.07DD525 pKa = 3.28SLYY528 pKa = 11.29NSMGYY533 pKa = 8.93IHH535 pKa = 7.53GKK537 pKa = 9.02PLIDD541 pKa = 4.58DD542 pKa = 4.39PRR544 pKa = 11.84WDD546 pKa = 3.27TGKK549 pKa = 9.33IHH551 pKa = 6.72VYY553 pKa = 8.5QWSIEE558 pKa = 4.17SGEE561 pKa = 4.11YY562 pKa = 8.66ATLVFEE568 pKa = 4.41VTDD571 pKa = 3.53YY572 pKa = 11.54SIGIEE577 pKa = 3.94KK578 pKa = 9.78LQSVEE583 pKa = 4.17TSPGLLDD590 pKa = 3.61GG591 pKa = 4.84
MM1 pKa = 7.42KK2 pKa = 10.67NKK4 pKa = 9.59IKK6 pKa = 10.68KK7 pKa = 9.93SFAAILALAMLLSLAACGGKK27 pKa = 10.14DD28 pKa = 3.62KK29 pKa = 11.42GSDD32 pKa = 3.38GDD34 pKa = 4.46GSDD37 pKa = 3.29KK38 pKa = 11.29TEE40 pKa = 4.16TIVLGKK46 pKa = 8.19YY47 pKa = 6.62TAVFKK52 pKa = 10.87GYY54 pKa = 8.09TLTKK58 pKa = 10.45DD59 pKa = 4.32DD60 pKa = 4.69EE61 pKa = 5.2GNDD64 pKa = 3.69AMVLTYY70 pKa = 10.51DD71 pKa = 3.64YY72 pKa = 11.25TNGSKK77 pKa = 10.69DD78 pKa = 3.55EE79 pKa = 4.25QSFAWAFLFEE89 pKa = 4.12ATQAGEE95 pKa = 4.14EE96 pKa = 4.62LGSPMYY102 pKa = 10.54DD103 pKa = 3.11EE104 pKa = 5.57NNNQITKK111 pKa = 10.44NYY113 pKa = 8.24EE114 pKa = 3.83EE115 pKa = 4.6KK116 pKa = 10.68VKK118 pKa = 10.53KK119 pKa = 10.52GQTLEE124 pKa = 3.89VAIALTLVNTTDD136 pKa = 3.32DD137 pKa = 4.15VIIVFSDD144 pKa = 3.77FDD146 pKa = 3.73DD147 pKa = 4.43HH148 pKa = 7.67EE149 pKa = 4.4YY150 pKa = 9.09TQTVKK155 pKa = 10.96LSGSGVQDD163 pKa = 3.34SGPEE167 pKa = 3.87EE168 pKa = 4.03AGSYY172 pKa = 8.63QLYY175 pKa = 10.6EE176 pKa = 3.9MTFEE180 pKa = 5.11GDD182 pKa = 3.69VFDD185 pKa = 5.78HH186 pKa = 7.46DD187 pKa = 4.23FLEE190 pKa = 4.65TSGFEE195 pKa = 4.36LYY197 pKa = 10.56TMEE200 pKa = 5.03LAADD204 pKa = 3.82GTGWLQIDD212 pKa = 3.98GRR214 pKa = 11.84GSLTWGDD221 pKa = 3.63GKK223 pKa = 9.47ITIDD227 pKa = 4.14GSDD230 pKa = 3.11AVYY233 pKa = 9.44TYY235 pKa = 9.56TYY237 pKa = 9.28KK238 pKa = 11.07DD239 pKa = 3.22GMIVLEE245 pKa = 4.25EE246 pKa = 4.21STGEE250 pKa = 3.98SMTFQKK256 pKa = 9.77TGAAGTSGGLKK267 pKa = 10.07PVGGDD272 pKa = 3.29GSDD275 pKa = 3.92ANLTAYY281 pKa = 8.18QQYY284 pKa = 10.37WNGDD288 pKa = 3.06WYY290 pKa = 11.17GWTAYY295 pKa = 11.21SNGTNDD301 pKa = 3.27YY302 pKa = 10.86SSLDD306 pKa = 3.86GFYY309 pKa = 8.99MDD311 pKa = 4.01CMARR315 pKa = 11.84IEE317 pKa = 4.01IDD319 pKa = 3.6EE320 pKa = 4.49NGEE323 pKa = 3.79GALILWDD330 pKa = 3.75EE331 pKa = 4.5ATSVDD336 pKa = 4.11DD337 pKa = 3.85PRR339 pKa = 11.84AGVTVCISDD348 pKa = 3.44GGFEE352 pKa = 3.95QGRR355 pKa = 11.84LKK357 pKa = 11.05SEE359 pKa = 4.83DD360 pKa = 2.96GWFSDD365 pKa = 4.16GDD367 pKa = 3.73VGNADD372 pKa = 3.62WLVDD376 pKa = 3.89PDD378 pKa = 3.79GLGYY382 pKa = 10.93EE383 pKa = 4.06NLICIEE389 pKa = 4.34GSYY392 pKa = 10.66KK393 pKa = 11.08DD394 pKa = 3.82PDD396 pKa = 4.19DD397 pKa = 4.34FFSGYY402 pKa = 10.79DD403 pKa = 3.22YY404 pKa = 10.96TIYY407 pKa = 10.54LRR409 pKa = 11.84PWGLDD414 pKa = 2.91WADD417 pKa = 3.82IEE419 pKa = 5.27ADD421 pKa = 3.83QPDD424 pKa = 4.96GLPDD428 pKa = 3.7HH429 pKa = 6.83YY430 pKa = 11.22DD431 pKa = 3.04WYY433 pKa = 11.25LSAIQTGAKK442 pKa = 8.52MPDD445 pKa = 3.92VIGGDD450 pKa = 3.84YY451 pKa = 11.07GGLAPGEE458 pKa = 4.41VPDD461 pKa = 5.16SQMEE465 pKa = 4.17PQGGNDD471 pKa = 3.7PNASAVDD478 pKa = 4.18GTWQAPVYY486 pKa = 10.67GDD488 pKa = 3.62YY489 pKa = 11.36GLSKK493 pKa = 10.91ASATGVVPMTRR504 pKa = 11.84DD505 pKa = 3.38EE506 pKa = 4.18LVDD509 pKa = 4.54RR510 pKa = 11.84IGFYY514 pKa = 10.97ADD516 pKa = 3.5SQIGSAPYY524 pKa = 10.07DD525 pKa = 3.28SLYY528 pKa = 11.29NSMGYY533 pKa = 8.93IHH535 pKa = 7.53GKK537 pKa = 9.02PLIDD541 pKa = 4.58DD542 pKa = 4.39PRR544 pKa = 11.84WDD546 pKa = 3.27TGKK549 pKa = 9.33IHH551 pKa = 6.72VYY553 pKa = 8.5QWSIEE558 pKa = 4.17SGEE561 pKa = 4.11YY562 pKa = 8.66ATLVFEE568 pKa = 4.41VTDD571 pKa = 3.53YY572 pKa = 11.54SIGIEE577 pKa = 3.94KK578 pKa = 9.78LQSVEE583 pKa = 4.17TSPGLLDD590 pKa = 3.61GG591 pKa = 4.84
Molecular weight: 64.36 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0S2W113|A0A0S2W113_9FIRM Uncharacterized protein OS=Intestinimonas butyriciproducens OX=1297617 GN=IB211_00606c PE=4 SV=1
MM1 pKa = 7.88VGILAAFSRR10 pKa = 11.84LRR12 pKa = 11.84LPWARR17 pKa = 11.84VSRR20 pKa = 11.84LRR22 pKa = 11.84RR23 pKa = 11.84GQFSQNIQKK32 pKa = 9.88FQGFVLL38 pKa = 3.98
MM1 pKa = 7.88VGILAAFSRR10 pKa = 11.84LRR12 pKa = 11.84LPWARR17 pKa = 11.84VSRR20 pKa = 11.84LRR22 pKa = 11.84RR23 pKa = 11.84GQFSQNIQKK32 pKa = 9.88FQGFVLL38 pKa = 3.98
Molecular weight: 4.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
986929 |
37 |
3097 |
293.9 |
32.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.003 ± 0.06 | 1.651 ± 0.019 |
5.504 ± 0.041 | 6.767 ± 0.048 |
3.779 ± 0.029 | 8.316 ± 0.045 |
1.793 ± 0.019 | 5.269 ± 0.041 |
4.352 ± 0.042 | 10.184 ± 0.064 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.757 ± 0.025 | 3.008 ± 0.029 |
4.388 ± 0.031 | 3.157 ± 0.027 |
6.096 ± 0.047 | 5.561 ± 0.043 |
5.565 ± 0.049 | 7.325 ± 0.034 |
1.085 ± 0.016 | 3.439 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
