
Velvet bean severe mosaic virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Begomovirus
Average proteome isoelectric point is 7.86
Get precalculated fractions of proteins
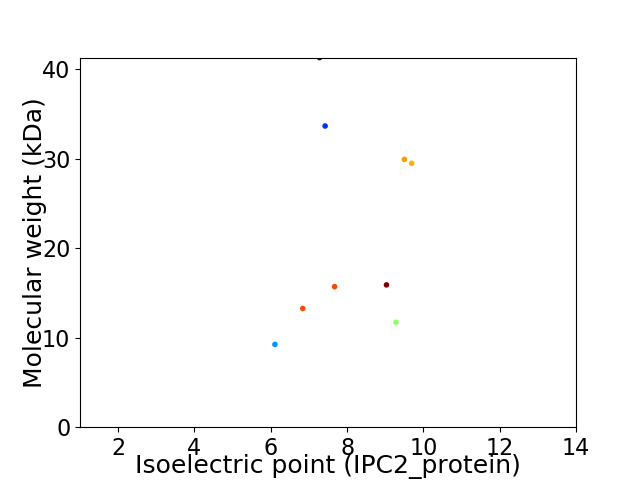
Virtual 2D-PAGE plot for 9 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
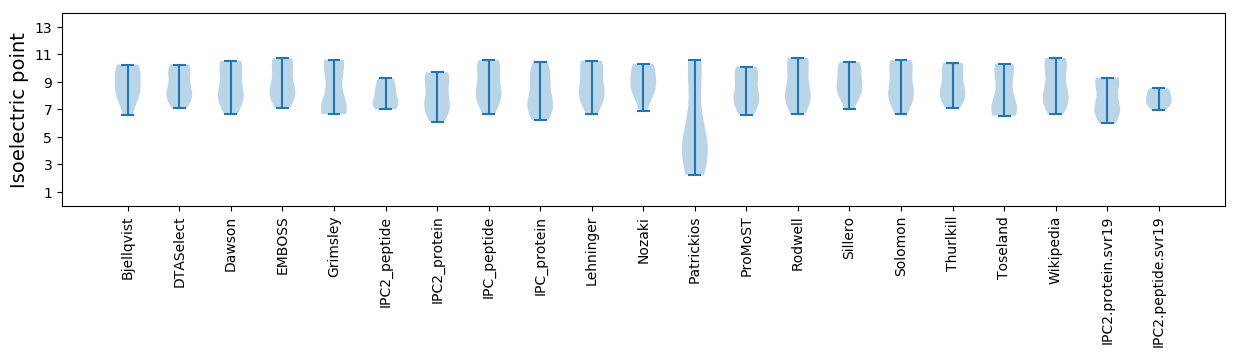
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C9YHF0|C9YHF0_9GEMI Replication enhancer OS=Velvet bean severe mosaic virus OX=667119 GN=AC3 PE=3 SV=1
MM1 pKa = 7.73VLILRR6 pKa = 11.84SFLVVIHH13 pKa = 6.0HH14 pKa = 6.67MIVNTEE20 pKa = 3.78VLVNHH25 pKa = 6.82GLLLTSILATCNSGIEE41 pKa = 4.08SSQYY45 pKa = 10.86LVTITKK51 pKa = 9.81IVFDD55 pKa = 4.13CCSAGFIIIHH65 pKa = 5.52VEE67 pKa = 3.79HH68 pKa = 6.59LTKK71 pKa = 10.21IHH73 pKa = 6.88RR74 pKa = 11.84CSEE77 pKa = 3.96WSSVTTT83 pKa = 4.07
MM1 pKa = 7.73VLILRR6 pKa = 11.84SFLVVIHH13 pKa = 6.0HH14 pKa = 6.67MIVNTEE20 pKa = 3.78VLVNHH25 pKa = 6.82GLLLTSILATCNSGIEE41 pKa = 4.08SSQYY45 pKa = 10.86LVTITKK51 pKa = 9.81IVFDD55 pKa = 4.13CCSAGFIIIHH65 pKa = 5.52VEE67 pKa = 3.79HH68 pKa = 6.59LTKK71 pKa = 10.21IHH73 pKa = 6.88RR74 pKa = 11.84CSEE77 pKa = 3.96WSSVTTT83 pKa = 4.07
Molecular weight: 9.25 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C9YHF5|C9YHF5_9GEMI Movement protein BC1 OS=Velvet bean severe mosaic virus OX=667119 GN=BC1 PE=3 SV=1
MM1 pKa = 7.43KK2 pKa = 9.75MLYY5 pKa = 9.16TPRR8 pKa = 11.84SNWRR12 pKa = 11.84SPFRR16 pKa = 11.84RR17 pKa = 11.84NTSNRR22 pKa = 11.84RR23 pKa = 11.84NVRR26 pKa = 11.84RR27 pKa = 11.84TNSATRR33 pKa = 11.84FNIKK37 pKa = 8.25VARR40 pKa = 11.84RR41 pKa = 11.84LSYY44 pKa = 10.99GRR46 pKa = 11.84VEE48 pKa = 4.6RR49 pKa = 11.84PLQFGKK55 pKa = 10.61LCEE58 pKa = 4.18KK59 pKa = 10.39HH60 pKa = 6.63HH61 pKa = 6.0GVHH64 pKa = 6.45MSLCSNRR71 pKa = 11.84DD72 pKa = 3.12VTSFISYY79 pKa = 8.18PALSLNGDD87 pKa = 3.21GRR89 pKa = 11.84SRR91 pKa = 11.84DD92 pKa = 4.09FIKK95 pKa = 10.82LLSLNISGVLSARR108 pKa = 11.84VVSSDD113 pKa = 3.16QPMDD117 pKa = 3.4NDD119 pKa = 3.58VYY121 pKa = 11.05RR122 pKa = 11.84RR123 pKa = 11.84GLFVISIILDD133 pKa = 3.72RR134 pKa = 11.84KK135 pKa = 10.07PYY137 pKa = 10.48VPDD140 pKa = 3.94GANEE144 pKa = 3.95LPSFEE149 pKa = 4.83EE150 pKa = 4.45LFGQYY155 pKa = 10.21SEE157 pKa = 6.06AYY159 pKa = 9.71VNMRR163 pKa = 11.84LLSSRR168 pKa = 11.84QDD170 pKa = 3.04RR171 pKa = 11.84FRR173 pKa = 11.84LLGTIKK179 pKa = 10.73KK180 pKa = 10.19NINCDD185 pKa = 3.09SGAADD190 pKa = 3.5VNIGKK195 pKa = 9.64FIRR198 pKa = 11.84FVQGRR203 pKa = 11.84RR204 pKa = 11.84TLWSRR209 pKa = 11.84FKK211 pKa = 11.11DD212 pKa = 3.58PEE214 pKa = 4.58PPLDD218 pKa = 3.53SGGNYY223 pKa = 10.08RR224 pKa = 11.84NISTNAILINYY235 pKa = 7.92AFVSMQSITVNPLVQYY251 pKa = 9.24EE252 pKa = 4.16LNYY255 pKa = 10.66VGG257 pKa = 5.28
MM1 pKa = 7.43KK2 pKa = 9.75MLYY5 pKa = 9.16TPRR8 pKa = 11.84SNWRR12 pKa = 11.84SPFRR16 pKa = 11.84RR17 pKa = 11.84NTSNRR22 pKa = 11.84RR23 pKa = 11.84NVRR26 pKa = 11.84RR27 pKa = 11.84TNSATRR33 pKa = 11.84FNIKK37 pKa = 8.25VARR40 pKa = 11.84RR41 pKa = 11.84LSYY44 pKa = 10.99GRR46 pKa = 11.84VEE48 pKa = 4.6RR49 pKa = 11.84PLQFGKK55 pKa = 10.61LCEE58 pKa = 4.18KK59 pKa = 10.39HH60 pKa = 6.63HH61 pKa = 6.0GVHH64 pKa = 6.45MSLCSNRR71 pKa = 11.84DD72 pKa = 3.12VTSFISYY79 pKa = 8.18PALSLNGDD87 pKa = 3.21GRR89 pKa = 11.84SRR91 pKa = 11.84DD92 pKa = 4.09FIKK95 pKa = 10.82LLSLNISGVLSARR108 pKa = 11.84VVSSDD113 pKa = 3.16QPMDD117 pKa = 3.4NDD119 pKa = 3.58VYY121 pKa = 11.05RR122 pKa = 11.84RR123 pKa = 11.84GLFVISIILDD133 pKa = 3.72RR134 pKa = 11.84KK135 pKa = 10.07PYY137 pKa = 10.48VPDD140 pKa = 3.94GANEE144 pKa = 3.95LPSFEE149 pKa = 4.83EE150 pKa = 4.45LFGQYY155 pKa = 10.21SEE157 pKa = 6.06AYY159 pKa = 9.71VNMRR163 pKa = 11.84LLSSRR168 pKa = 11.84QDD170 pKa = 3.04RR171 pKa = 11.84FRR173 pKa = 11.84LLGTIKK179 pKa = 10.73KK180 pKa = 10.19NINCDD185 pKa = 3.09SGAADD190 pKa = 3.5VNIGKK195 pKa = 9.64FIRR198 pKa = 11.84FVQGRR203 pKa = 11.84RR204 pKa = 11.84TLWSRR209 pKa = 11.84FKK211 pKa = 11.11DD212 pKa = 3.58PEE214 pKa = 4.58PPLDD218 pKa = 3.53SGGNYY223 pKa = 10.08RR224 pKa = 11.84NISTNAILINYY235 pKa = 7.92AFVSMQSITVNPLVQYY251 pKa = 9.24EE252 pKa = 4.16LNYY255 pKa = 10.66VGG257 pKa = 5.28
Molecular weight: 29.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1741 |
83 |
362 |
193.4 |
22.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.055 ± 0.788 | 2.24 ± 0.391 |
4.825 ± 0.517 | 5.055 ± 0.583 |
4.882 ± 0.178 | 4.48 ± 0.459 |
2.929 ± 0.489 | 5.859 ± 0.659 |
6.203 ± 0.788 | 7.812 ± 0.863 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.585 ± 0.532 | 6.72 ± 0.482 |
4.767 ± 0.547 | 3.906 ± 0.409 |
7.122 ± 0.989 | 8.558 ± 0.869 |
5.859 ± 0.749 | 6.031 ± 0.828 |
1.091 ± 0.088 | 4.021 ± 0.548 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
