
Flectobacillus sp. BAB-3569
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Cytophagaceae; Flectobacillus; unclassified Flectobacillus
Average proteome isoelectric point is 6.78
Get precalculated fractions of proteins
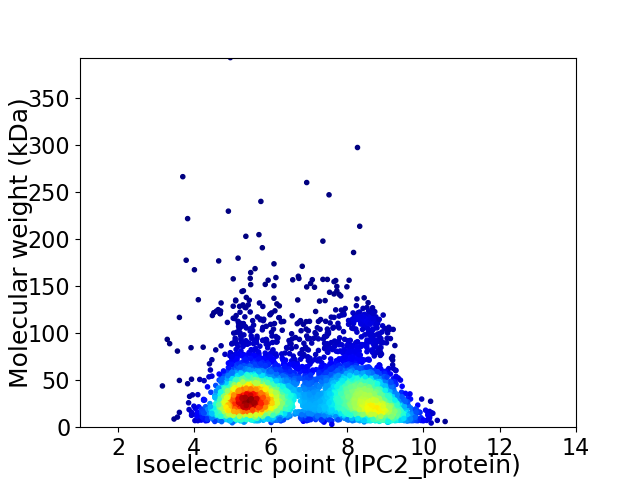
Virtual 2D-PAGE plot for 5084 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A267T4J3|A0A267T4J3_9BACT Ornithine--oxo-acid aminotransferase OS=Flectobacillus sp. BAB-3569 OX=1509483 GN=rocD PE=3 SV=1
MM1 pKa = 7.68NILQKK6 pKa = 10.39IVILLVCLTLVSVKK20 pKa = 10.09IQAQDD25 pKa = 3.2VSINIINQPSTVTQGSTLGRR45 pKa = 11.84VTVDD49 pKa = 2.6ICNNDD54 pKa = 3.17GGSRR58 pKa = 11.84SAAANRR64 pKa = 11.84LRR66 pKa = 11.84PLITLPSSLVGSVIPITTTGWTVLPIDD93 pKa = 4.09INNPTTLRR101 pKa = 11.84FEE103 pKa = 4.24NTVAIAPGEE112 pKa = 4.03CSQIVIGYY120 pKa = 7.34TGVNIGGPLTVTGTLGFNGPQTTGNLTGNDD150 pKa = 3.45NSTTSITVITGDD162 pKa = 3.54TDD164 pKa = 3.97GDD166 pKa = 4.09GDD168 pKa = 4.8PDD170 pKa = 3.5ITDD173 pKa = 4.08PQPTDD178 pKa = 3.04PCAWGTGQVLANTTTAWRR196 pKa = 11.84TADD199 pKa = 3.7CDD201 pKa = 3.85GDD203 pKa = 4.25GVTNYY208 pKa = 10.36KK209 pKa = 10.12EE210 pKa = 4.07ATGTDD215 pKa = 4.16DD216 pKa = 6.2DD217 pKa = 5.17PLTLGDD223 pKa = 3.9NTNPNDD229 pKa = 3.62GCSYY233 pKa = 10.94NKK235 pKa = 9.61VDD237 pKa = 3.69QVIGNVSASWKK248 pKa = 9.17TLDD251 pKa = 4.56CDD253 pKa = 4.38KK254 pKa = 11.21DD255 pKa = 3.92GNPNEE260 pKa = 4.45TDD262 pKa = 3.94LNPQVATAVNDD273 pKa = 3.36VLTAPFGLTSTVSVLSNDD291 pKa = 3.36DD292 pKa = 3.94FLPVASNEE300 pKa = 3.61ITRR303 pKa = 11.84IGGNASGTVAFAPTTGVMSYY323 pKa = 9.35TPSATEE329 pKa = 3.6PGGPNVTVVYY339 pKa = 7.71QVCNTLVTPNVCASATVTISVPAAGDD365 pKa = 3.31QDD367 pKa = 4.59GDD369 pKa = 3.86GDD371 pKa = 4.8PDD373 pKa = 3.69NTDD376 pKa = 3.45PQPTNSCVWGIGQVLANTSTTWRR399 pKa = 11.84NADD402 pKa = 3.47CDD404 pKa = 4.03GDD406 pKa = 4.45GVTNYY411 pKa = 10.62NEE413 pKa = 3.83ATGADD418 pKa = 4.14GNPATLADD426 pKa = 3.81NTDD429 pKa = 3.83PLSACSLNLGQVXIRR444 pKa = 11.84FVLLLDD450 pKa = 3.46YY451 pKa = 11.35SGFLRR456 pKa = 11.84NFANSLDD463 pKa = 3.88TPLILLSGQIQYY475 pKa = 11.52GKK477 pKa = 9.52MFFGG481 pKa = 4.32
MM1 pKa = 7.68NILQKK6 pKa = 10.39IVILLVCLTLVSVKK20 pKa = 10.09IQAQDD25 pKa = 3.2VSINIINQPSTVTQGSTLGRR45 pKa = 11.84VTVDD49 pKa = 2.6ICNNDD54 pKa = 3.17GGSRR58 pKa = 11.84SAAANRR64 pKa = 11.84LRR66 pKa = 11.84PLITLPSSLVGSVIPITTTGWTVLPIDD93 pKa = 4.09INNPTTLRR101 pKa = 11.84FEE103 pKa = 4.24NTVAIAPGEE112 pKa = 4.03CSQIVIGYY120 pKa = 7.34TGVNIGGPLTVTGTLGFNGPQTTGNLTGNDD150 pKa = 3.45NSTTSITVITGDD162 pKa = 3.54TDD164 pKa = 3.97GDD166 pKa = 4.09GDD168 pKa = 4.8PDD170 pKa = 3.5ITDD173 pKa = 4.08PQPTDD178 pKa = 3.04PCAWGTGQVLANTTTAWRR196 pKa = 11.84TADD199 pKa = 3.7CDD201 pKa = 3.85GDD203 pKa = 4.25GVTNYY208 pKa = 10.36KK209 pKa = 10.12EE210 pKa = 4.07ATGTDD215 pKa = 4.16DD216 pKa = 6.2DD217 pKa = 5.17PLTLGDD223 pKa = 3.9NTNPNDD229 pKa = 3.62GCSYY233 pKa = 10.94NKK235 pKa = 9.61VDD237 pKa = 3.69QVIGNVSASWKK248 pKa = 9.17TLDD251 pKa = 4.56CDD253 pKa = 4.38KK254 pKa = 11.21DD255 pKa = 3.92GNPNEE260 pKa = 4.45TDD262 pKa = 3.94LNPQVATAVNDD273 pKa = 3.36VLTAPFGLTSTVSVLSNDD291 pKa = 3.36DD292 pKa = 3.94FLPVASNEE300 pKa = 3.61ITRR303 pKa = 11.84IGGNASGTVAFAPTTGVMSYY323 pKa = 9.35TPSATEE329 pKa = 3.6PGGPNVTVVYY339 pKa = 7.71QVCNTLVTPNVCASATVTISVPAAGDD365 pKa = 3.31QDD367 pKa = 4.59GDD369 pKa = 3.86GDD371 pKa = 4.8PDD373 pKa = 3.69NTDD376 pKa = 3.45PQPTNSCVWGIGQVLANTSTTWRR399 pKa = 11.84NADD402 pKa = 3.47CDD404 pKa = 4.03GDD406 pKa = 4.45GVTNYY411 pKa = 10.62NEE413 pKa = 3.83ATGADD418 pKa = 4.14GNPATLADD426 pKa = 3.81NTDD429 pKa = 3.83PLSACSLNLGQVXIRR444 pKa = 11.84FVLLLDD450 pKa = 3.46YY451 pKa = 11.35SGFLRR456 pKa = 11.84NFANSLDD463 pKa = 3.88TPLILLSGQIQYY475 pKa = 11.52GKK477 pKa = 9.52MFFGG481 pKa = 4.32
Molecular weight: 49.72 kDa
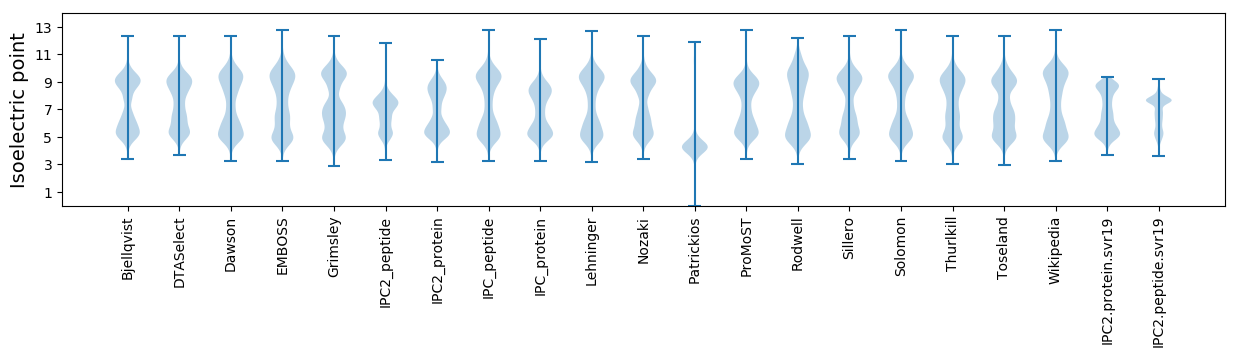
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A267T511|A0A267T511_9BACT NAGPA domain-containing protein OS=Flectobacillus sp. BAB-3569 OX=1509483 GN=BWI92_20980 PE=4 SV=1
MM1 pKa = 7.57KK2 pKa = 10.25KK3 pKa = 9.95VEE5 pKa = 4.0YY6 pKa = 10.32LNRR9 pKa = 11.84RR10 pKa = 11.84MRR12 pKa = 11.84LKK14 pKa = 10.33IVSDD18 pKa = 3.97DD19 pKa = 3.65FLTISTLEE27 pKa = 3.84YY28 pKa = 10.29RR29 pKa = 11.84VGSFLGIGIWRR40 pKa = 11.84RR41 pKa = 11.84VGDD44 pKa = 3.81RR45 pKa = 11.84VEE47 pKa = 4.76LGSNVQLQQIVLSIQFTRR65 pKa = 11.84RR66 pKa = 11.84KK67 pKa = 9.95NKK69 pKa = 9.91LEE71 pKa = 3.78RR72 pKa = 11.84RR73 pKa = 11.84ASLTQIAEE81 pKa = 4.49KK82 pKa = 10.38LCKK85 pKa = 9.28TATPP89 pKa = 4.06
MM1 pKa = 7.57KK2 pKa = 10.25KK3 pKa = 9.95VEE5 pKa = 4.0YY6 pKa = 10.32LNRR9 pKa = 11.84RR10 pKa = 11.84MRR12 pKa = 11.84LKK14 pKa = 10.33IVSDD18 pKa = 3.97DD19 pKa = 3.65FLTISTLEE27 pKa = 3.84YY28 pKa = 10.29RR29 pKa = 11.84VGSFLGIGIWRR40 pKa = 11.84RR41 pKa = 11.84VGDD44 pKa = 3.81RR45 pKa = 11.84VEE47 pKa = 4.76LGSNVQLQQIVLSIQFTRR65 pKa = 11.84RR66 pKa = 11.84KK67 pKa = 9.95NKK69 pKa = 9.91LEE71 pKa = 3.78RR72 pKa = 11.84RR73 pKa = 11.84ASLTQIAEE81 pKa = 4.49KK82 pKa = 10.38LCKK85 pKa = 9.28TATPP89 pKa = 4.06
Molecular weight: 10.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1755997 |
26 |
3834 |
345.4 |
38.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.918 ± 0.036 | 0.838 ± 0.011 |
5.082 ± 0.023 | 6.035 ± 0.045 |
4.962 ± 0.025 | 6.611 ± 0.036 |
1.791 ± 0.018 | 7.137 ± 0.03 |
7.206 ± 0.038 | 9.519 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.2 ± 0.018 | 5.602 ± 0.036 |
3.751 ± 0.02 | 4.147 ± 0.024 |
3.678 ± 0.022 | 6.755 ± 0.038 |
5.946 ± 0.043 | 6.435 ± 0.031 |
1.293 ± 0.015 | 4.093 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
