
Bat coronavirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Cornidovirineae; Coronaviridae; Coronavirinae; unclassified Coronavirinae
Average proteome isoelectric point is 7.01
Get precalculated fractions of proteins
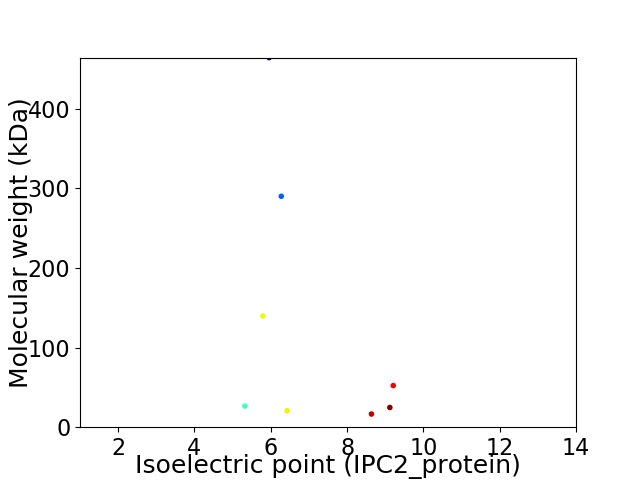
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Z4EVQ5|A0A2Z4EVQ5_9NIDO ORF3 OS=Bat coronavirus OX=1508220 PE=4 SV=1
MM1 pKa = 7.22MLINLLRR8 pKa = 11.84KK9 pKa = 10.05LYY11 pKa = 10.14GSPAAPQQAISHH23 pKa = 6.55DD24 pKa = 3.81VSIVNAEE31 pKa = 3.95ALEE34 pKa = 4.1PVYY37 pKa = 11.27NMPEE41 pKa = 3.87VEE43 pKa = 4.35ANEE46 pKa = 4.11SLLLFILGSVFYY58 pKa = 10.95VLLSLILWLLSTVNKK73 pKa = 9.58PYY75 pKa = 10.88LSIVSSVIFCFVQLVMVIWFVLCDD99 pKa = 3.58NVLISIVAQCFLAFLFVMCFLEE121 pKa = 4.17RR122 pKa = 11.84SYY124 pKa = 11.55LAFRR128 pKa = 11.84LRR130 pKa = 11.84SLAPFVHH137 pKa = 6.28SCSAFVVINTTSNRR151 pKa = 11.84YY152 pKa = 8.32VFPLQGVAEE161 pKa = 4.28HH162 pKa = 6.36VVILTTNAGVSCNGVFQQGAIAVTDD187 pKa = 3.33TATIYY192 pKa = 10.52TRR194 pKa = 11.84FSSHH198 pKa = 6.12VLLLDD203 pKa = 3.96RR204 pKa = 11.84VEE206 pKa = 4.62HH207 pKa = 6.45GYY209 pKa = 11.03DD210 pKa = 3.18YY211 pKa = 11.3TVFAFVSTVVLEE223 pKa = 3.87NTRR226 pKa = 11.84RR227 pKa = 11.84IGVSTGMTDD236 pKa = 3.36VEE238 pKa = 4.44LL239 pKa = 4.93
MM1 pKa = 7.22MLINLLRR8 pKa = 11.84KK9 pKa = 10.05LYY11 pKa = 10.14GSPAAPQQAISHH23 pKa = 6.55DD24 pKa = 3.81VSIVNAEE31 pKa = 3.95ALEE34 pKa = 4.1PVYY37 pKa = 11.27NMPEE41 pKa = 3.87VEE43 pKa = 4.35ANEE46 pKa = 4.11SLLLFILGSVFYY58 pKa = 10.95VLLSLILWLLSTVNKK73 pKa = 9.58PYY75 pKa = 10.88LSIVSSVIFCFVQLVMVIWFVLCDD99 pKa = 3.58NVLISIVAQCFLAFLFVMCFLEE121 pKa = 4.17RR122 pKa = 11.84SYY124 pKa = 11.55LAFRR128 pKa = 11.84LRR130 pKa = 11.84SLAPFVHH137 pKa = 6.28SCSAFVVINTTSNRR151 pKa = 11.84YY152 pKa = 8.32VFPLQGVAEE161 pKa = 4.28HH162 pKa = 6.36VVILTTNAGVSCNGVFQQGAIAVTDD187 pKa = 3.33TATIYY192 pKa = 10.52TRR194 pKa = 11.84FSSHH198 pKa = 6.12VLLLDD203 pKa = 3.96RR204 pKa = 11.84VEE206 pKa = 4.62HH207 pKa = 6.45GYY209 pKa = 11.03DD210 pKa = 3.18YY211 pKa = 11.3TVFAFVSTVVLEE223 pKa = 3.87NTRR226 pKa = 11.84RR227 pKa = 11.84IGVSTGMTDD236 pKa = 3.36VEE238 pKa = 4.44LL239 pKa = 4.93
Molecular weight: 26.58 kDa
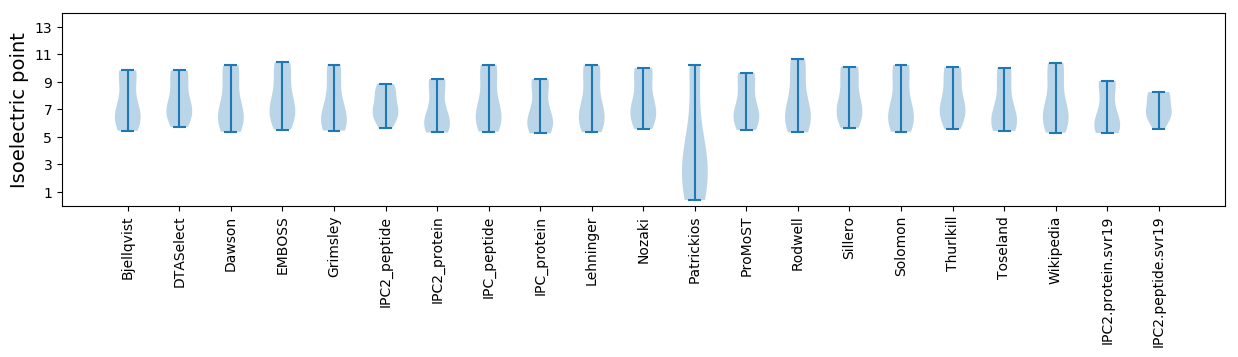
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z4EVP6|A0A2Z4EVP6_9NIDO Uncharacterized protein OS=Bat coronavirus OX=1508220 PE=4 SV=1
MM1 pKa = 7.42SGRR4 pKa = 11.84RR5 pKa = 11.84TPRR8 pKa = 11.84NQPQVSFKK16 pKa = 11.16NEE18 pKa = 3.78SDD20 pKa = 3.53SDD22 pKa = 4.25SEE24 pKa = 4.34SGARR28 pKa = 11.84SQSRR32 pKa = 11.84GRR34 pKa = 11.84NSNNNNNGGNGGARR48 pKa = 11.84RR49 pKa = 11.84KK50 pKa = 10.1DD51 pKa = 3.45KK52 pKa = 10.77PEE54 pKa = 3.97KK55 pKa = 9.96PRR57 pKa = 11.84AAPAQNVSWFLPIVQTGKK75 pKa = 9.96QDD77 pKa = 3.19LRR79 pKa = 11.84FARR82 pKa = 11.84GEE84 pKa = 4.16GVPVSQGVDD93 pKa = 2.6ITYY96 pKa = 10.22QHH98 pKa = 6.81GYY100 pKa = 6.01WLRR103 pKa = 11.84RR104 pKa = 11.84QRR106 pKa = 11.84TFNKK110 pKa = 10.11GGKK113 pKa = 7.95QVQANPRR120 pKa = 11.84WFFYY124 pKa = 9.86YY125 pKa = 10.12TGTGPYY131 pKa = 9.53EE132 pKa = 3.84GLRR135 pKa = 11.84YY136 pKa = 9.92GSRR139 pKa = 11.84NNDD142 pKa = 3.71IIWVGNEE149 pKa = 3.46GANVNRR155 pKa = 11.84LGDD158 pKa = 3.38MGTRR162 pKa = 11.84NPANDD167 pKa = 3.39AGIPVQLAEE176 pKa = 5.13GIPKK180 pKa = 10.24GFYY183 pKa = 10.51AEE185 pKa = 4.29GRR187 pKa = 11.84NSRR190 pKa = 11.84GNSRR194 pKa = 11.84NSSRR198 pKa = 11.84SSSRR202 pKa = 11.84GSSNANSRR210 pKa = 11.84NQSRR214 pKa = 11.84SNSPGRR220 pKa = 11.84GSAPPSGGEE229 pKa = 3.31PWMAYY234 pKa = 10.38LIQKK238 pKa = 10.09LEE240 pKa = 4.0NLEE243 pKa = 3.82QRR245 pKa = 11.84VDD247 pKa = 3.43GKK249 pKa = 11.17KK250 pKa = 9.94SDD252 pKa = 3.66KK253 pKa = 10.99QPVKK257 pKa = 8.99VTKK260 pKa = 10.61NVASEE265 pKa = 3.86NAKK268 pKa = 10.25KK269 pKa = 10.25LRR271 pKa = 11.84HH272 pKa = 6.07KK273 pKa = 9.87RR274 pKa = 11.84TAHH277 pKa = 6.33KK278 pKa = 10.56GSNATQNYY286 pKa = 7.8GRR288 pKa = 11.84RR289 pKa = 11.84GPGNLEE295 pKa = 4.0GNFGDD300 pKa = 5.09QEE302 pKa = 4.17FLKK305 pKa = 10.93LGTDD309 pKa = 3.63DD310 pKa = 3.63PRR312 pKa = 11.84FPVVAQMAPNTSSFVFMSHH331 pKa = 5.45FTPRR335 pKa = 11.84YY336 pKa = 8.26EE337 pKa = 6.0ADD339 pKa = 4.35ALWLDD344 pKa = 3.67YY345 pKa = 10.64TGSIKK350 pKa = 10.65LPRR353 pKa = 11.84DD354 pKa = 3.78DD355 pKa = 5.33PNFPQWEE362 pKa = 4.24KK363 pKa = 11.29LLAEE367 pKa = 5.1NIDD370 pKa = 3.92AYY372 pKa = 10.3KK373 pKa = 10.54SFPPPKK379 pKa = 9.59PKK381 pKa = 9.89SDD383 pKa = 3.29KK384 pKa = 10.4KK385 pKa = 10.84KK386 pKa = 10.84KK387 pKa = 9.75SDD389 pKa = 3.95KK390 pKa = 10.35SDD392 pKa = 3.34SAAGPSEE399 pKa = 4.28DD400 pKa = 4.39LQMQVVDD407 pKa = 3.55PSGVQRR413 pKa = 11.84IYY415 pKa = 10.71MKK417 pKa = 10.71DD418 pKa = 3.22AADD421 pKa = 3.57QTDD424 pKa = 4.45DD425 pKa = 3.93EE426 pKa = 4.65WLQDD430 pKa = 3.35DD431 pKa = 5.4TIYY434 pKa = 11.08EE435 pKa = 4.23DD436 pKa = 5.13EE437 pKa = 4.1NDD439 pKa = 3.5KK440 pKa = 11.22PKK442 pKa = 10.71AQRR445 pKa = 11.84RR446 pKa = 11.84QSIKK450 pKa = 10.28KK451 pKa = 10.2RR452 pKa = 11.84NATHH456 pKa = 5.6QRR458 pKa = 11.84HH459 pKa = 4.8VSIDD463 pKa = 3.25GAAQSSAA470 pKa = 3.23
MM1 pKa = 7.42SGRR4 pKa = 11.84RR5 pKa = 11.84TPRR8 pKa = 11.84NQPQVSFKK16 pKa = 11.16NEE18 pKa = 3.78SDD20 pKa = 3.53SDD22 pKa = 4.25SEE24 pKa = 4.34SGARR28 pKa = 11.84SQSRR32 pKa = 11.84GRR34 pKa = 11.84NSNNNNNGGNGGARR48 pKa = 11.84RR49 pKa = 11.84KK50 pKa = 10.1DD51 pKa = 3.45KK52 pKa = 10.77PEE54 pKa = 3.97KK55 pKa = 9.96PRR57 pKa = 11.84AAPAQNVSWFLPIVQTGKK75 pKa = 9.96QDD77 pKa = 3.19LRR79 pKa = 11.84FARR82 pKa = 11.84GEE84 pKa = 4.16GVPVSQGVDD93 pKa = 2.6ITYY96 pKa = 10.22QHH98 pKa = 6.81GYY100 pKa = 6.01WLRR103 pKa = 11.84RR104 pKa = 11.84QRR106 pKa = 11.84TFNKK110 pKa = 10.11GGKK113 pKa = 7.95QVQANPRR120 pKa = 11.84WFFYY124 pKa = 9.86YY125 pKa = 10.12TGTGPYY131 pKa = 9.53EE132 pKa = 3.84GLRR135 pKa = 11.84YY136 pKa = 9.92GSRR139 pKa = 11.84NNDD142 pKa = 3.71IIWVGNEE149 pKa = 3.46GANVNRR155 pKa = 11.84LGDD158 pKa = 3.38MGTRR162 pKa = 11.84NPANDD167 pKa = 3.39AGIPVQLAEE176 pKa = 5.13GIPKK180 pKa = 10.24GFYY183 pKa = 10.51AEE185 pKa = 4.29GRR187 pKa = 11.84NSRR190 pKa = 11.84GNSRR194 pKa = 11.84NSSRR198 pKa = 11.84SSSRR202 pKa = 11.84GSSNANSRR210 pKa = 11.84NQSRR214 pKa = 11.84SNSPGRR220 pKa = 11.84GSAPPSGGEE229 pKa = 3.31PWMAYY234 pKa = 10.38LIQKK238 pKa = 10.09LEE240 pKa = 4.0NLEE243 pKa = 3.82QRR245 pKa = 11.84VDD247 pKa = 3.43GKK249 pKa = 11.17KK250 pKa = 9.94SDD252 pKa = 3.66KK253 pKa = 10.99QPVKK257 pKa = 8.99VTKK260 pKa = 10.61NVASEE265 pKa = 3.86NAKK268 pKa = 10.25KK269 pKa = 10.25LRR271 pKa = 11.84HH272 pKa = 6.07KK273 pKa = 9.87RR274 pKa = 11.84TAHH277 pKa = 6.33KK278 pKa = 10.56GSNATQNYY286 pKa = 7.8GRR288 pKa = 11.84RR289 pKa = 11.84GPGNLEE295 pKa = 4.0GNFGDD300 pKa = 5.09QEE302 pKa = 4.17FLKK305 pKa = 10.93LGTDD309 pKa = 3.63DD310 pKa = 3.63PRR312 pKa = 11.84FPVVAQMAPNTSSFVFMSHH331 pKa = 5.45FTPRR335 pKa = 11.84YY336 pKa = 8.26EE337 pKa = 6.0ADD339 pKa = 4.35ALWLDD344 pKa = 3.67YY345 pKa = 10.64TGSIKK350 pKa = 10.65LPRR353 pKa = 11.84DD354 pKa = 3.78DD355 pKa = 5.33PNFPQWEE362 pKa = 4.24KK363 pKa = 11.29LLAEE367 pKa = 5.1NIDD370 pKa = 3.92AYY372 pKa = 10.3KK373 pKa = 10.54SFPPPKK379 pKa = 9.59PKK381 pKa = 9.89SDD383 pKa = 3.29KK384 pKa = 10.4KK385 pKa = 10.84KK386 pKa = 10.84KK387 pKa = 9.75SDD389 pKa = 3.95KK390 pKa = 10.35SDD392 pKa = 3.34SAAGPSEE399 pKa = 4.28DD400 pKa = 4.39LQMQVVDD407 pKa = 3.55PSGVQRR413 pKa = 11.84IYY415 pKa = 10.71MKK417 pKa = 10.71DD418 pKa = 3.22AADD421 pKa = 3.57QTDD424 pKa = 4.45DD425 pKa = 3.93EE426 pKa = 4.65WLQDD430 pKa = 3.35DD431 pKa = 5.4TIYY434 pKa = 11.08EE435 pKa = 4.23DD436 pKa = 5.13EE437 pKa = 4.1NDD439 pKa = 3.5KK440 pKa = 11.22PKK442 pKa = 10.71AQRR445 pKa = 11.84RR446 pKa = 11.84QSIKK450 pKa = 10.28KK451 pKa = 10.2RR452 pKa = 11.84NATHH456 pKa = 5.6QRR458 pKa = 11.84HH459 pKa = 4.8VSIDD463 pKa = 3.25GAAQSSAA470 pKa = 3.23
Molecular weight: 52.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
9311 |
146 |
4205 |
1163.9 |
129.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.421 ± 0.357 | 2.9 ± 0.312 |
5.241 ± 0.551 | 3.684 ± 0.344 |
5.069 ± 0.386 | 6.036 ± 0.322 |
1.751 ± 0.262 | 4.339 ± 0.249 |
4.704 ± 0.608 | 9.344 ± 0.485 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.255 ± 0.215 | 5.091 ± 0.551 |
4.672 ± 0.276 | 3.566 ± 0.395 |
4.189 ± 0.412 | 7.239 ± 0.674 |
6.927 ± 0.537 | 9.612 ± 0.54 |
1.31 ± 0.138 | 4.65 ± 0.291 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
