
Aestuariispira insulae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Kiloniellaceae; Aestuariispira
Average proteome isoelectric point is 5.99
Get precalculated fractions of proteins
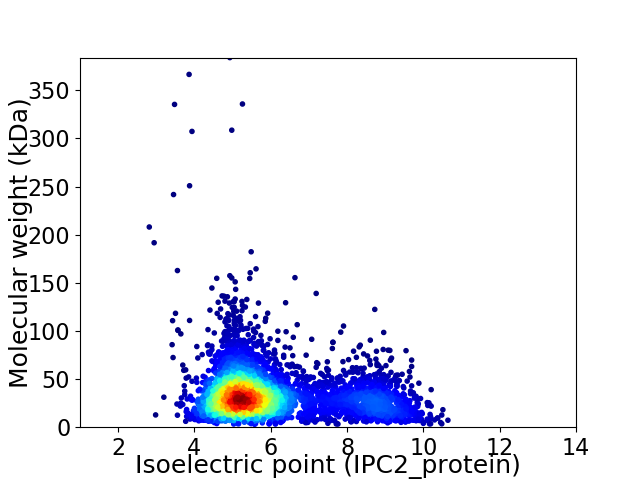
Virtual 2D-PAGE plot for 4490 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
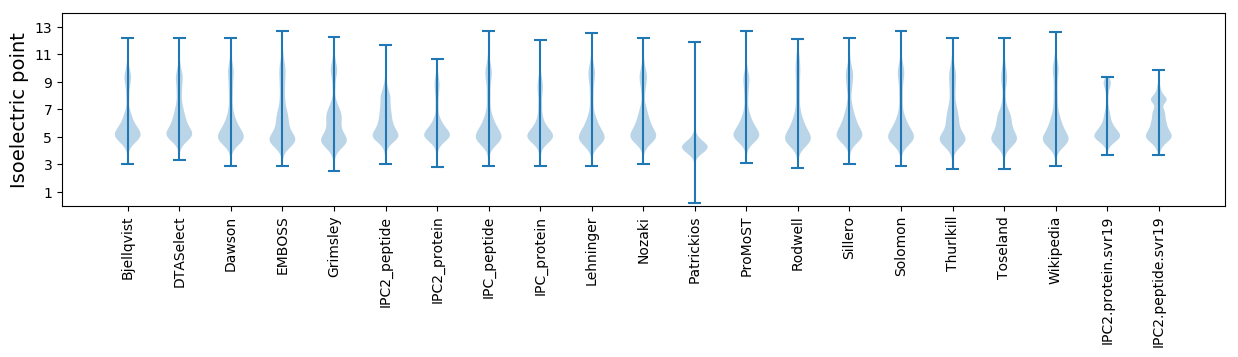
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3D9H5Q5|A0A3D9H5Q5_9PROT Choline monooxygenase OS=Aestuariispira insulae OX=1461337 GN=DFP90_11323 PE=4 SV=1
MM1 pKa = 7.62SSLNSALSAAVSGLFAQSTSISAISTNLSNVGTTAYY37 pKa = 9.85KK38 pKa = 9.45EE39 pKa = 4.05ASVSFSTLVSGSTGNNRR56 pKa = 11.84SSNFNGSGVYY66 pKa = 10.32AGVNQSISHH75 pKa = 7.23LGNVSDD81 pKa = 4.29TAISTNIALTGSGFLVVSDD100 pKa = 4.73DD101 pKa = 3.7AAGDD105 pKa = 3.46EE106 pKa = 5.18LYY108 pKa = 10.54FSRR111 pKa = 11.84VGSFYY116 pKa = 10.12PDD118 pKa = 3.46KK119 pKa = 10.81DD120 pKa = 3.78DD121 pKa = 5.83KK122 pKa = 11.4LVNLNGFYY130 pKa = 10.78LQGYY134 pKa = 6.07PTDD137 pKa = 3.72ASGNPTVGTGSTSTLEE153 pKa = 4.38TIDD156 pKa = 4.07LGSIGGNAAATEE168 pKa = 4.49NIDD171 pKa = 2.96IRR173 pKa = 11.84ANIPANAAIGDD184 pKa = 4.12SFNTDD189 pKa = 2.06IEE191 pKa = 4.53MFDD194 pKa = 3.6SLGTSHH200 pKa = 6.86YY201 pKa = 9.73VTATWTKK208 pKa = 8.89TAANTWDD215 pKa = 3.15ITYY218 pKa = 10.57ADD220 pKa = 4.45PVLSADD226 pKa = 3.54STTTTGTVTGSSTITFDD243 pKa = 3.23SDD245 pKa = 3.78GNFVSSTSNNLAINGWTTGASNSAITMNFAGLTQFTKK282 pKa = 10.77DD283 pKa = 3.42EE284 pKa = 4.02QTLDD288 pKa = 3.56IEE290 pKa = 4.33IDD292 pKa = 3.86KK293 pKa = 10.89INQDD297 pKa = 3.05GLEE300 pKa = 4.02YY301 pKa = 11.28GEE303 pKa = 4.94LKK305 pKa = 10.64AVSIGSDD312 pKa = 3.5GLVTATFEE320 pKa = 4.4NGTSYY325 pKa = 10.7PVYY328 pKa = 9.74QIPVATFSNPNGLALEE344 pKa = 4.43SGNVYY349 pKa = 10.6SATNDD354 pKa = 3.02SGTYY358 pKa = 8.41TLHH361 pKa = 7.72DD362 pKa = 5.16PGDD365 pKa = 4.08GGAGEE370 pKa = 4.72LKK372 pKa = 10.65GSALEE377 pKa = 4.31GSTVDD382 pKa = 4.55TADD385 pKa = 3.46QLTKK389 pKa = 10.94LIVAQQAYY397 pKa = 8.45SACAEE402 pKa = 4.23IVSTTDD408 pKa = 4.17DD409 pKa = 3.72MFDD412 pKa = 3.62TLISAARR419 pKa = 3.5
MM1 pKa = 7.62SSLNSALSAAVSGLFAQSTSISAISTNLSNVGTTAYY37 pKa = 9.85KK38 pKa = 9.45EE39 pKa = 4.05ASVSFSTLVSGSTGNNRR56 pKa = 11.84SSNFNGSGVYY66 pKa = 10.32AGVNQSISHH75 pKa = 7.23LGNVSDD81 pKa = 4.29TAISTNIALTGSGFLVVSDD100 pKa = 4.73DD101 pKa = 3.7AAGDD105 pKa = 3.46EE106 pKa = 5.18LYY108 pKa = 10.54FSRR111 pKa = 11.84VGSFYY116 pKa = 10.12PDD118 pKa = 3.46KK119 pKa = 10.81DD120 pKa = 3.78DD121 pKa = 5.83KK122 pKa = 11.4LVNLNGFYY130 pKa = 10.78LQGYY134 pKa = 6.07PTDD137 pKa = 3.72ASGNPTVGTGSTSTLEE153 pKa = 4.38TIDD156 pKa = 4.07LGSIGGNAAATEE168 pKa = 4.49NIDD171 pKa = 2.96IRR173 pKa = 11.84ANIPANAAIGDD184 pKa = 4.12SFNTDD189 pKa = 2.06IEE191 pKa = 4.53MFDD194 pKa = 3.6SLGTSHH200 pKa = 6.86YY201 pKa = 9.73VTATWTKK208 pKa = 8.89TAANTWDD215 pKa = 3.15ITYY218 pKa = 10.57ADD220 pKa = 4.45PVLSADD226 pKa = 3.54STTTTGTVTGSSTITFDD243 pKa = 3.23SDD245 pKa = 3.78GNFVSSTSNNLAINGWTTGASNSAITMNFAGLTQFTKK282 pKa = 10.77DD283 pKa = 3.42EE284 pKa = 4.02QTLDD288 pKa = 3.56IEE290 pKa = 4.33IDD292 pKa = 3.86KK293 pKa = 10.89INQDD297 pKa = 3.05GLEE300 pKa = 4.02YY301 pKa = 11.28GEE303 pKa = 4.94LKK305 pKa = 10.64AVSIGSDD312 pKa = 3.5GLVTATFEE320 pKa = 4.4NGTSYY325 pKa = 10.7PVYY328 pKa = 9.74QIPVATFSNPNGLALEE344 pKa = 4.43SGNVYY349 pKa = 10.6SATNDD354 pKa = 3.02SGTYY358 pKa = 8.41TLHH361 pKa = 7.72DD362 pKa = 5.16PGDD365 pKa = 4.08GGAGEE370 pKa = 4.72LKK372 pKa = 10.65GSALEE377 pKa = 4.31GSTVDD382 pKa = 4.55TADD385 pKa = 3.46QLTKK389 pKa = 10.94LIVAQQAYY397 pKa = 8.45SACAEE402 pKa = 4.23IVSTTDD408 pKa = 4.17DD409 pKa = 3.72MFDD412 pKa = 3.62TLISAARR419 pKa = 3.5
Molecular weight: 43.09 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3D9HK64|A0A3D9HK64_9PROT CelD/BcsL family acetyltransferase involved in cellulose biosynthesis OS=Aestuariispira insulae OX=1461337 GN=DFP90_105261 PE=4 SV=1
MM1 pKa = 7.77EE2 pKa = 5.66FGTWVIYY9 pKa = 10.36LFACAALCFTPGPNSLLALSHH30 pKa = 5.9GASYY34 pKa = 10.86GFKK37 pKa = 9.62RR38 pKa = 11.84TIFTSAGGALGFAAVIAVCLAGLGALLAASATAFLVIKK76 pKa = 10.29LVGAAYY82 pKa = 9.62LIWLGIQSLRR92 pKa = 11.84RR93 pKa = 11.84PAFVLGADD101 pKa = 3.75VAAKK105 pKa = 9.52GLRR108 pKa = 11.84RR109 pKa = 11.84RR110 pKa = 11.84SHH112 pKa = 6.65FAQGFLAAATNPKK125 pKa = 10.29GILFFVAFLPQFYY138 pKa = 10.72DD139 pKa = 3.44PARR142 pKa = 11.84PAFLQFLILAATFASIEE159 pKa = 4.13FFLEE163 pKa = 3.69LALAALAHH171 pKa = 7.15RR172 pKa = 11.84LRR174 pKa = 11.84PWLAKK179 pKa = 10.51ASVGKK184 pKa = 9.21WFKK187 pKa = 10.64RR188 pKa = 11.84VTGASFIGIGAGLATASGRR207 pKa = 3.73
MM1 pKa = 7.77EE2 pKa = 5.66FGTWVIYY9 pKa = 10.36LFACAALCFTPGPNSLLALSHH30 pKa = 5.9GASYY34 pKa = 10.86GFKK37 pKa = 9.62RR38 pKa = 11.84TIFTSAGGALGFAAVIAVCLAGLGALLAASATAFLVIKK76 pKa = 10.29LVGAAYY82 pKa = 9.62LIWLGIQSLRR92 pKa = 11.84RR93 pKa = 11.84PAFVLGADD101 pKa = 3.75VAAKK105 pKa = 9.52GLRR108 pKa = 11.84RR109 pKa = 11.84RR110 pKa = 11.84SHH112 pKa = 6.65FAQGFLAAATNPKK125 pKa = 10.29GILFFVAFLPQFYY138 pKa = 10.72DD139 pKa = 3.44PARR142 pKa = 11.84PAFLQFLILAATFASIEE159 pKa = 4.13FFLEE163 pKa = 3.69LALAALAHH171 pKa = 7.15RR172 pKa = 11.84LRR174 pKa = 11.84PWLAKK179 pKa = 10.51ASVGKK184 pKa = 9.21WFKK187 pKa = 10.64RR188 pKa = 11.84VTGASFIGIGAGLATASGRR207 pKa = 3.73
Molecular weight: 21.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1443071 |
28 |
3550 |
321.4 |
35.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.236 ± 0.044 | 0.975 ± 0.013 |
6.237 ± 0.042 | 6.487 ± 0.034 |
3.885 ± 0.027 | 8.185 ± 0.045 |
2.176 ± 0.021 | 5.726 ± 0.024 |
4.171 ± 0.031 | 10.307 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.817 ± 0.019 | 3.241 ± 0.024 |
4.62 ± 0.027 | 3.633 ± 0.026 |
6.113 ± 0.037 | 5.654 ± 0.031 |
5.028 ± 0.032 | 6.749 ± 0.035 |
1.271 ± 0.015 | 2.489 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
