
Gammapapillomavirus 7
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
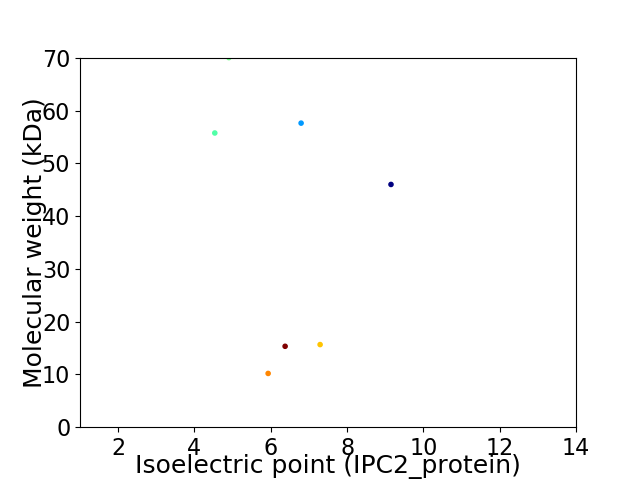
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2D2ALC5|A0A2D2ALC5_9PAPI Regulatory protein E2 OS=Gammapapillomavirus 7 OX=1175849 GN=E2 PE=3 SV=1
MM1 pKa = 8.1DD2 pKa = 3.86SEE4 pKa = 4.64RR5 pKa = 11.84PRR7 pKa = 11.84KK8 pKa = 8.02RR9 pKa = 11.84TKK11 pKa = 10.15RR12 pKa = 11.84DD13 pKa = 3.08SVTNLYY19 pKa = 7.64RR20 pKa = 11.84TCQISGNCPEE30 pKa = 4.16DD31 pKa = 3.32VKK33 pKa = 11.62NKK35 pKa = 10.04VEE37 pKa = 4.34ANTLADD43 pKa = 3.34KK44 pKa = 10.86LLRR47 pKa = 11.84ILSSIVYY54 pKa = 9.96FGGLGIGTGKK64 pKa = 8.78GTGGSTGYY72 pKa = 10.49RR73 pKa = 11.84PISSGGGGGGRR84 pKa = 11.84VTGDD88 pKa = 3.05GTVIRR93 pKa = 11.84PNIVVEE99 pKa = 4.07PVGPSDD105 pKa = 5.19VIPIDD110 pKa = 3.61ALSPGSSSIVPLQEE124 pKa = 3.95AGAEE128 pKa = 4.07IIIPDD133 pKa = 3.52ISTDD137 pKa = 3.39LGAGEE142 pKa = 5.07LEE144 pKa = 4.97VITEE148 pKa = 4.14PDD150 pKa = 3.51IIDD153 pKa = 3.05ISGPQEE159 pKa = 4.21SPTVTTSEE167 pKa = 4.87DD168 pKa = 2.89IAIIDD173 pKa = 3.97VQPGPSTPRR182 pKa = 11.84RR183 pKa = 11.84VVTSSSRR190 pKa = 11.84FSNPSFISVVTSSSTTEE207 pKa = 3.99AGSTAIDD214 pKa = 3.87VFVHH218 pKa = 6.25GAAGGDD224 pKa = 3.56IIGEE228 pKa = 4.38EE229 pKa = 4.06IPLDD233 pKa = 3.58TFNEE237 pKa = 4.12PEE239 pKa = 3.97EE240 pKa = 4.42FEE242 pKa = 5.27IEE244 pKa = 5.33DD245 pKa = 3.54IPQPRR250 pKa = 11.84SSTPRR255 pKa = 11.84AFQRR259 pKa = 11.84AFSRR263 pKa = 11.84ARR265 pKa = 11.84DD266 pKa = 3.56LYY268 pKa = 9.85NRR270 pKa = 11.84RR271 pKa = 11.84VRR273 pKa = 11.84QVMTQNINFLTRR285 pKa = 11.84APQAVQFAFEE295 pKa = 4.96NPAFDD300 pKa = 3.77NDD302 pKa = 3.21ITIEE306 pKa = 4.05FQQDD310 pKa = 3.35LNQLAAAAPDD320 pKa = 4.13PDD322 pKa = 3.6FADD325 pKa = 3.05IVRR328 pKa = 11.84LHH330 pKa = 6.86RR331 pKa = 11.84PRR333 pKa = 11.84FSEE336 pKa = 3.96TAEE339 pKa = 3.84GQIRR343 pKa = 11.84VSRR346 pKa = 11.84LGTKK350 pKa = 8.53GTIRR354 pKa = 11.84LRR356 pKa = 11.84SGTQIGEE363 pKa = 4.49TVHH366 pKa = 7.0FYY368 pKa = 11.54YY369 pKa = 10.63DD370 pKa = 3.62LSSIEE375 pKa = 4.0DD376 pKa = 3.42AEE378 pKa = 4.58AIEE381 pKa = 4.44LSVLGEE387 pKa = 4.03HH388 pKa = 6.72SGDD391 pKa = 3.1ATVINPQAEE400 pKa = 4.59STFVDD405 pKa = 4.38AEE407 pKa = 4.12NSDD410 pKa = 3.86APLLFPEE417 pKa = 5.06EE418 pKa = 4.34EE419 pKa = 4.16LLDD422 pKa = 5.32DD423 pKa = 4.48ISEE426 pKa = 4.58DD427 pKa = 3.66FSNSHH432 pKa = 6.94VILSSGGRR440 pKa = 11.84RR441 pKa = 11.84SVLSVPTLPPGVALKK456 pKa = 10.81VFIDD460 pKa = 4.05DD461 pKa = 3.45VGEE464 pKa = 4.1GLFVSHH470 pKa = 7.11PVSTDD475 pKa = 3.34SIPPGVIPISDD486 pKa = 3.86IQPSILIDD494 pKa = 3.59EE495 pKa = 4.96FSSDD499 pKa = 4.99DD500 pKa = 4.21FILHH504 pKa = 7.02PSHH507 pKa = 6.25IPKK510 pKa = 10.32RR511 pKa = 11.84KK512 pKa = 8.36RR513 pKa = 11.84KK514 pKa = 9.46RR515 pKa = 11.84VTSLL519 pKa = 2.88
MM1 pKa = 8.1DD2 pKa = 3.86SEE4 pKa = 4.64RR5 pKa = 11.84PRR7 pKa = 11.84KK8 pKa = 8.02RR9 pKa = 11.84TKK11 pKa = 10.15RR12 pKa = 11.84DD13 pKa = 3.08SVTNLYY19 pKa = 7.64RR20 pKa = 11.84TCQISGNCPEE30 pKa = 4.16DD31 pKa = 3.32VKK33 pKa = 11.62NKK35 pKa = 10.04VEE37 pKa = 4.34ANTLADD43 pKa = 3.34KK44 pKa = 10.86LLRR47 pKa = 11.84ILSSIVYY54 pKa = 9.96FGGLGIGTGKK64 pKa = 8.78GTGGSTGYY72 pKa = 10.49RR73 pKa = 11.84PISSGGGGGGRR84 pKa = 11.84VTGDD88 pKa = 3.05GTVIRR93 pKa = 11.84PNIVVEE99 pKa = 4.07PVGPSDD105 pKa = 5.19VIPIDD110 pKa = 3.61ALSPGSSSIVPLQEE124 pKa = 3.95AGAEE128 pKa = 4.07IIIPDD133 pKa = 3.52ISTDD137 pKa = 3.39LGAGEE142 pKa = 5.07LEE144 pKa = 4.97VITEE148 pKa = 4.14PDD150 pKa = 3.51IIDD153 pKa = 3.05ISGPQEE159 pKa = 4.21SPTVTTSEE167 pKa = 4.87DD168 pKa = 2.89IAIIDD173 pKa = 3.97VQPGPSTPRR182 pKa = 11.84RR183 pKa = 11.84VVTSSSRR190 pKa = 11.84FSNPSFISVVTSSSTTEE207 pKa = 3.99AGSTAIDD214 pKa = 3.87VFVHH218 pKa = 6.25GAAGGDD224 pKa = 3.56IIGEE228 pKa = 4.38EE229 pKa = 4.06IPLDD233 pKa = 3.58TFNEE237 pKa = 4.12PEE239 pKa = 3.97EE240 pKa = 4.42FEE242 pKa = 5.27IEE244 pKa = 5.33DD245 pKa = 3.54IPQPRR250 pKa = 11.84SSTPRR255 pKa = 11.84AFQRR259 pKa = 11.84AFSRR263 pKa = 11.84ARR265 pKa = 11.84DD266 pKa = 3.56LYY268 pKa = 9.85NRR270 pKa = 11.84RR271 pKa = 11.84VRR273 pKa = 11.84QVMTQNINFLTRR285 pKa = 11.84APQAVQFAFEE295 pKa = 4.96NPAFDD300 pKa = 3.77NDD302 pKa = 3.21ITIEE306 pKa = 4.05FQQDD310 pKa = 3.35LNQLAAAAPDD320 pKa = 4.13PDD322 pKa = 3.6FADD325 pKa = 3.05IVRR328 pKa = 11.84LHH330 pKa = 6.86RR331 pKa = 11.84PRR333 pKa = 11.84FSEE336 pKa = 3.96TAEE339 pKa = 3.84GQIRR343 pKa = 11.84VSRR346 pKa = 11.84LGTKK350 pKa = 8.53GTIRR354 pKa = 11.84LRR356 pKa = 11.84SGTQIGEE363 pKa = 4.49TVHH366 pKa = 7.0FYY368 pKa = 11.54YY369 pKa = 10.63DD370 pKa = 3.62LSSIEE375 pKa = 4.0DD376 pKa = 3.42AEE378 pKa = 4.58AIEE381 pKa = 4.44LSVLGEE387 pKa = 4.03HH388 pKa = 6.72SGDD391 pKa = 3.1ATVINPQAEE400 pKa = 4.59STFVDD405 pKa = 4.38AEE407 pKa = 4.12NSDD410 pKa = 3.86APLLFPEE417 pKa = 5.06EE418 pKa = 4.34EE419 pKa = 4.16LLDD422 pKa = 5.32DD423 pKa = 4.48ISEE426 pKa = 4.58DD427 pKa = 3.66FSNSHH432 pKa = 6.94VILSSGGRR440 pKa = 11.84RR441 pKa = 11.84SVLSVPTLPPGVALKK456 pKa = 10.81VFIDD460 pKa = 4.05DD461 pKa = 3.45VGEE464 pKa = 4.1GLFVSHH470 pKa = 7.11PVSTDD475 pKa = 3.34SIPPGVIPISDD486 pKa = 3.86IQPSILIDD494 pKa = 3.59EE495 pKa = 4.96FSSDD499 pKa = 4.99DD500 pKa = 4.21FILHH504 pKa = 7.02PSHH507 pKa = 6.25IPKK510 pKa = 10.32RR511 pKa = 11.84KK512 pKa = 8.36RR513 pKa = 11.84KK514 pKa = 9.46RR515 pKa = 11.84VTSLL519 pKa = 2.88
Molecular weight: 55.75 kDa
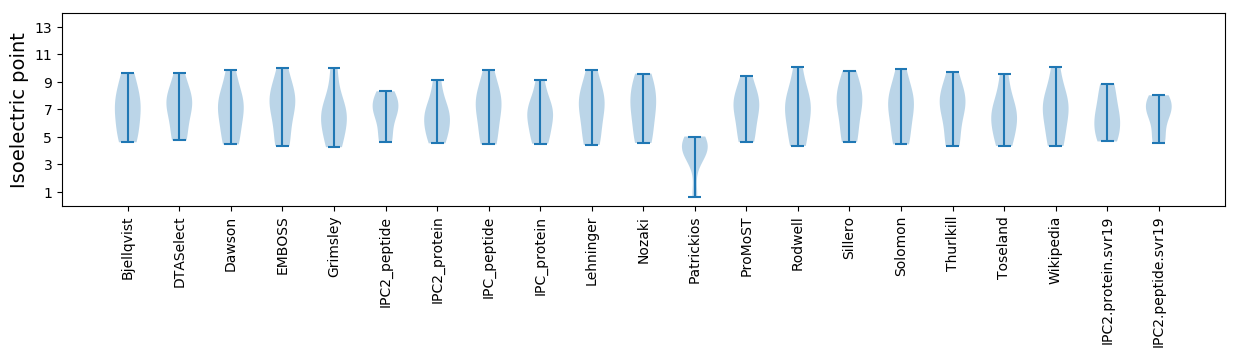
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2D2ALC6|A0A2D2ALC6_9PAPI ^E4 (Fragment) OS=Gammapapillomavirus 7 OX=1175849 GN=^E4 PE=4 SV=1
MM1 pKa = 7.57EE2 pKa = 4.83SLAEE6 pKa = 4.43RR7 pKa = 11.84FDD9 pKa = 4.13ALQEE13 pKa = 4.18NQLTLYY19 pKa = 7.69EE20 pKa = 4.3TGSTNIEE27 pKa = 3.92DD28 pKa = 4.48QILYY32 pKa = 9.33WDD34 pKa = 4.05TVRR37 pKa = 11.84QEE39 pKa = 4.14YY40 pKa = 11.14VLFHH44 pKa = 5.95YY45 pKa = 10.3ARR47 pKa = 11.84QRR49 pKa = 11.84GLSHH53 pKa = 7.28IGLQPVPTLTVSEE66 pKa = 4.51QKK68 pKa = 10.75AKK70 pKa = 10.47QAILMVLQLKK80 pKa = 8.89SLKK83 pKa = 9.98QSQFGNEE90 pKa = 3.95PWTMQDD96 pKa = 2.44TSYY99 pKa = 11.41EE100 pKa = 4.2LFNTAPSNTFKK111 pKa = 10.85KK112 pKa = 10.15GAYY115 pKa = 8.18TVDD118 pKa = 3.05VYY120 pKa = 11.9YY121 pKa = 11.01DD122 pKa = 3.8GDD124 pKa = 3.6EE125 pKa = 4.67DD126 pKa = 5.07NYY128 pKa = 10.74FPYY131 pKa = 10.0TAWSYY136 pKa = 11.28IYY138 pKa = 9.86YY139 pKa = 10.45QNGDD143 pKa = 3.96NEE145 pKa = 4.16WHH147 pKa = 6.37KK148 pKa = 11.48VKK150 pKa = 10.95GDD152 pKa = 3.1IDD154 pKa = 4.34YY155 pKa = 11.13EE156 pKa = 4.35GLFYY160 pKa = 10.77KK161 pKa = 10.49SHH163 pKa = 7.33DD164 pKa = 3.84GEE166 pKa = 4.05KK167 pKa = 9.84VYY169 pKa = 11.25YY170 pKa = 8.49VTFYY174 pKa = 11.59DD175 pKa = 3.64DD176 pKa = 3.19ATRR179 pKa = 11.84FSRR182 pKa = 11.84TGKK185 pKa = 6.65WTVRR189 pKa = 11.84YY190 pKa = 9.85KK191 pKa = 10.16NTTISSTSITSTSGPTGNPTSKK213 pKa = 10.23PSSDD217 pKa = 3.45SVGQKK222 pKa = 8.88ATEE225 pKa = 3.89QASKK229 pKa = 10.49RR230 pKa = 11.84RR231 pKa = 11.84QRR233 pKa = 11.84SPSPSSPRR241 pKa = 11.84HH242 pKa = 4.34TRR244 pKa = 11.84SRR246 pKa = 11.84TWQSDD251 pKa = 2.78GDD253 pKa = 4.18TSSSSPGISTRR264 pKa = 11.84RR265 pKa = 11.84RR266 pKa = 11.84RR267 pKa = 11.84RR268 pKa = 11.84EE269 pKa = 3.98GKK271 pKa = 8.32STTQRR276 pKa = 11.84RR277 pKa = 11.84EE278 pKa = 3.75RR279 pKa = 11.84PSRR282 pKa = 11.84ATSPGVAPEE291 pKa = 4.0EE292 pKa = 3.93VGRR295 pKa = 11.84RR296 pKa = 11.84HH297 pKa = 6.98RR298 pKa = 11.84FTQRR302 pKa = 11.84KK303 pKa = 7.77GLSRR307 pKa = 11.84LGQLQEE313 pKa = 4.32DD314 pKa = 4.14ARR316 pKa = 11.84DD317 pKa = 3.63PPIILLKK324 pKa = 10.96GPANTLKK331 pKa = 10.57CYY333 pKa = 10.45RR334 pKa = 11.84FRR336 pKa = 11.84CRR338 pKa = 11.84SKK340 pKa = 10.62CRR342 pKa = 11.84SLIYY346 pKa = 10.52RR347 pKa = 11.84MSTVFSWVGEE357 pKa = 4.22GPEE360 pKa = 3.94RR361 pKa = 11.84LGEE364 pKa = 4.03PRR366 pKa = 11.84MLLAFKK372 pKa = 10.59DD373 pKa = 3.39KK374 pKa = 10.34EE375 pKa = 3.78QRR377 pKa = 11.84QRR379 pKa = 11.84FLSTVHH385 pKa = 6.13LPKK388 pKa = 9.91GTHH391 pKa = 5.62YY392 pKa = 11.42SLGNLNSLL400 pKa = 4.21
MM1 pKa = 7.57EE2 pKa = 4.83SLAEE6 pKa = 4.43RR7 pKa = 11.84FDD9 pKa = 4.13ALQEE13 pKa = 4.18NQLTLYY19 pKa = 7.69EE20 pKa = 4.3TGSTNIEE27 pKa = 3.92DD28 pKa = 4.48QILYY32 pKa = 9.33WDD34 pKa = 4.05TVRR37 pKa = 11.84QEE39 pKa = 4.14YY40 pKa = 11.14VLFHH44 pKa = 5.95YY45 pKa = 10.3ARR47 pKa = 11.84QRR49 pKa = 11.84GLSHH53 pKa = 7.28IGLQPVPTLTVSEE66 pKa = 4.51QKK68 pKa = 10.75AKK70 pKa = 10.47QAILMVLQLKK80 pKa = 8.89SLKK83 pKa = 9.98QSQFGNEE90 pKa = 3.95PWTMQDD96 pKa = 2.44TSYY99 pKa = 11.41EE100 pKa = 4.2LFNTAPSNTFKK111 pKa = 10.85KK112 pKa = 10.15GAYY115 pKa = 8.18TVDD118 pKa = 3.05VYY120 pKa = 11.9YY121 pKa = 11.01DD122 pKa = 3.8GDD124 pKa = 3.6EE125 pKa = 4.67DD126 pKa = 5.07NYY128 pKa = 10.74FPYY131 pKa = 10.0TAWSYY136 pKa = 11.28IYY138 pKa = 9.86YY139 pKa = 10.45QNGDD143 pKa = 3.96NEE145 pKa = 4.16WHH147 pKa = 6.37KK148 pKa = 11.48VKK150 pKa = 10.95GDD152 pKa = 3.1IDD154 pKa = 4.34YY155 pKa = 11.13EE156 pKa = 4.35GLFYY160 pKa = 10.77KK161 pKa = 10.49SHH163 pKa = 7.33DD164 pKa = 3.84GEE166 pKa = 4.05KK167 pKa = 9.84VYY169 pKa = 11.25YY170 pKa = 8.49VTFYY174 pKa = 11.59DD175 pKa = 3.64DD176 pKa = 3.19ATRR179 pKa = 11.84FSRR182 pKa = 11.84TGKK185 pKa = 6.65WTVRR189 pKa = 11.84YY190 pKa = 9.85KK191 pKa = 10.16NTTISSTSITSTSGPTGNPTSKK213 pKa = 10.23PSSDD217 pKa = 3.45SVGQKK222 pKa = 8.88ATEE225 pKa = 3.89QASKK229 pKa = 10.49RR230 pKa = 11.84RR231 pKa = 11.84QRR233 pKa = 11.84SPSPSSPRR241 pKa = 11.84HH242 pKa = 4.34TRR244 pKa = 11.84SRR246 pKa = 11.84TWQSDD251 pKa = 2.78GDD253 pKa = 4.18TSSSSPGISTRR264 pKa = 11.84RR265 pKa = 11.84RR266 pKa = 11.84RR267 pKa = 11.84RR268 pKa = 11.84EE269 pKa = 3.98GKK271 pKa = 8.32STTQRR276 pKa = 11.84RR277 pKa = 11.84EE278 pKa = 3.75RR279 pKa = 11.84PSRR282 pKa = 11.84ATSPGVAPEE291 pKa = 4.0EE292 pKa = 3.93VGRR295 pKa = 11.84RR296 pKa = 11.84HH297 pKa = 6.98RR298 pKa = 11.84FTQRR302 pKa = 11.84KK303 pKa = 7.77GLSRR307 pKa = 11.84LGQLQEE313 pKa = 4.32DD314 pKa = 4.14ARR316 pKa = 11.84DD317 pKa = 3.63PPIILLKK324 pKa = 10.96GPANTLKK331 pKa = 10.57CYY333 pKa = 10.45RR334 pKa = 11.84FRR336 pKa = 11.84CRR338 pKa = 11.84SKK340 pKa = 10.62CRR342 pKa = 11.84SLIYY346 pKa = 10.52RR347 pKa = 11.84MSTVFSWVGEE357 pKa = 4.22GPEE360 pKa = 3.94RR361 pKa = 11.84LGEE364 pKa = 4.03PRR366 pKa = 11.84MLLAFKK372 pKa = 10.59DD373 pKa = 3.39KK374 pKa = 10.34EE375 pKa = 3.78QRR377 pKa = 11.84QRR379 pKa = 11.84FLSTVHH385 pKa = 6.13LPKK388 pKa = 9.91GTHH391 pKa = 5.62YY392 pKa = 11.42SLGNLNSLL400 pKa = 4.21
Molecular weight: 46.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2413 |
93 |
617 |
344.7 |
38.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.263 ± 0.438 | 2.196 ± 0.788 |
6.672 ± 0.469 | 5.802 ± 0.783 |
4.31 ± 0.443 | 6.009 ± 0.792 |
1.782 ± 0.117 | 5.636 ± 0.945 |
5.222 ± 0.792 | 8.91 ± 0.848 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.575 ± 0.374 | 4.642 ± 0.729 |
6.051 ± 0.964 | 4.434 ± 0.371 |
5.802 ± 1.05 | 8.869 ± 0.792 |
6.299 ± 0.695 | 6.216 ± 0.573 |
1.202 ± 0.304 | 3.108 ± 0.634 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
