
Allonocardiopsis opalescens
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptosporangiales; Streptosporangiales incertae sedis; Allonocardiopsis
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
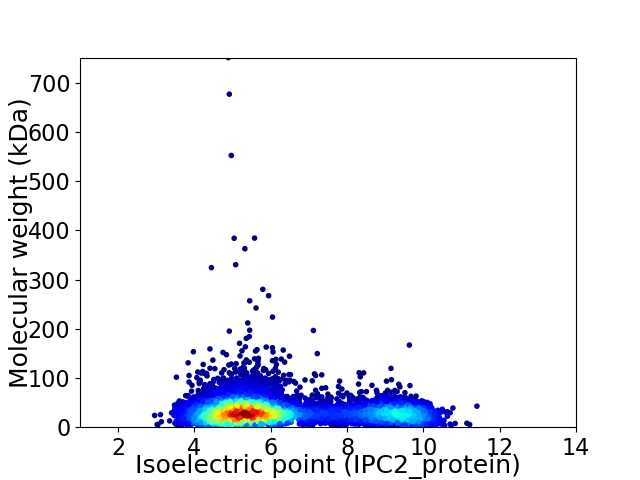
Virtual 2D-PAGE plot for 6300 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2T0PVL4|A0A2T0PVL4_9ACTN Uncharacterized protein OS=Allonocardiopsis opalescens OX=1144618 GN=CLV72_109195 PE=4 SV=1
MM1 pKa = 7.57KK2 pKa = 10.04IIHH5 pKa = 6.61ALIAVLAVAALTACAPLGAPPVADD29 pKa = 4.79LPPTEE34 pKa = 4.62PLADD38 pKa = 3.76EE39 pKa = 4.52ATVDD43 pKa = 3.37AAEE46 pKa = 4.35AAEE49 pKa = 4.4SPVEE53 pKa = 4.01EE54 pKa = 5.21GPAEE58 pKa = 4.28AEE60 pKa = 4.22LGSTHH65 pKa = 5.76TWNDD69 pKa = 2.7GLAIEE74 pKa = 4.53LSGVEE79 pKa = 3.79RR80 pKa = 11.84ALSGEE85 pKa = 4.13YY86 pKa = 10.68AMPAGEE92 pKa = 4.15PYY94 pKa = 10.85VRR96 pKa = 11.84FTMQVQNDD104 pKa = 3.56TGGPVDD110 pKa = 5.64LSLVWIEE117 pKa = 4.27CQVGEE122 pKa = 4.69DD123 pKa = 3.8GRR125 pKa = 11.84IGDD128 pKa = 4.54LVYY131 pKa = 10.68DD132 pKa = 4.06YY133 pKa = 11.48DD134 pKa = 5.46AGLGDD139 pKa = 4.36GFSSTVMDD147 pKa = 4.66GRR149 pKa = 11.84DD150 pKa = 3.16ATADD154 pKa = 3.98FGCAMPEE161 pKa = 4.0DD162 pKa = 4.2EE163 pKa = 6.28SYY165 pKa = 11.32LQIEE169 pKa = 4.36VHH171 pKa = 6.16PQDD174 pKa = 3.58EE175 pKa = 4.49AMLRR179 pKa = 11.84QPVFFIGDD187 pKa = 3.56VPP189 pKa = 3.78
MM1 pKa = 7.57KK2 pKa = 10.04IIHH5 pKa = 6.61ALIAVLAVAALTACAPLGAPPVADD29 pKa = 4.79LPPTEE34 pKa = 4.62PLADD38 pKa = 3.76EE39 pKa = 4.52ATVDD43 pKa = 3.37AAEE46 pKa = 4.35AAEE49 pKa = 4.4SPVEE53 pKa = 4.01EE54 pKa = 5.21GPAEE58 pKa = 4.28AEE60 pKa = 4.22LGSTHH65 pKa = 5.76TWNDD69 pKa = 2.7GLAIEE74 pKa = 4.53LSGVEE79 pKa = 3.79RR80 pKa = 11.84ALSGEE85 pKa = 4.13YY86 pKa = 10.68AMPAGEE92 pKa = 4.15PYY94 pKa = 10.85VRR96 pKa = 11.84FTMQVQNDD104 pKa = 3.56TGGPVDD110 pKa = 5.64LSLVWIEE117 pKa = 4.27CQVGEE122 pKa = 4.69DD123 pKa = 3.8GRR125 pKa = 11.84IGDD128 pKa = 4.54LVYY131 pKa = 10.68DD132 pKa = 4.06YY133 pKa = 11.48DD134 pKa = 5.46AGLGDD139 pKa = 4.36GFSSTVMDD147 pKa = 4.66GRR149 pKa = 11.84DD150 pKa = 3.16ATADD154 pKa = 3.98FGCAMPEE161 pKa = 4.0DD162 pKa = 4.2EE163 pKa = 6.28SYY165 pKa = 11.32LQIEE169 pKa = 4.36VHH171 pKa = 6.16PQDD174 pKa = 3.58EE175 pKa = 4.49AMLRR179 pKa = 11.84QPVFFIGDD187 pKa = 3.56VPP189 pKa = 3.78
Molecular weight: 19.88 kDa
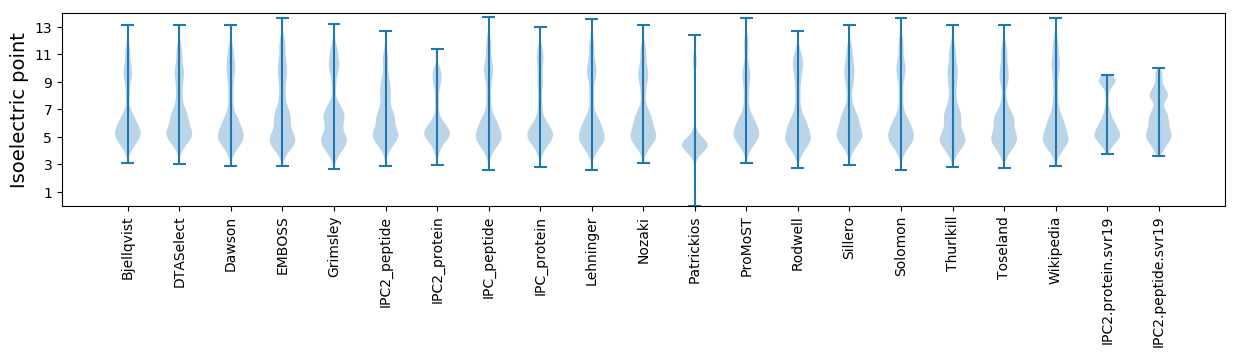
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2T0PU41|A0A2T0PU41_9ACTN Trk system potassium uptake protein TrkA OS=Allonocardiopsis opalescens OX=1144618 GN=CLV72_110179 PE=4 SV=1
MM1 pKa = 7.34NPAIHH6 pKa = 6.77RR7 pKa = 11.84RR8 pKa = 11.84HH9 pKa = 5.56TPALIPNSAPTPPKK23 pKa = 7.0TTPRR27 pKa = 11.84ASQQVAGIHH36 pKa = 5.21RR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84TRR42 pKa = 11.84PRR44 pKa = 11.84HH45 pKa = 4.87HH46 pKa = 6.97LTRR49 pKa = 11.84IPRR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84LHH56 pKa = 5.53PTRR59 pKa = 11.84LTRR62 pKa = 11.84PRR64 pKa = 11.84RR65 pKa = 11.84RR66 pKa = 11.84RR67 pKa = 11.84RR68 pKa = 11.84PRR70 pKa = 11.84LPTTPRR76 pKa = 11.84RR77 pKa = 11.84TRR79 pKa = 11.84RR80 pKa = 11.84PPTRR84 pKa = 11.84RR85 pKa = 11.84RR86 pKa = 11.84RR87 pKa = 11.84RR88 pKa = 11.84RR89 pKa = 11.84RR90 pKa = 11.84RR91 pKa = 11.84NRR93 pKa = 11.84RR94 pKa = 11.84RR95 pKa = 11.84RR96 pKa = 11.84RR97 pKa = 11.84HH98 pKa = 4.61LRR100 pKa = 11.84LRR102 pKa = 11.84RR103 pKa = 11.84HH104 pKa = 6.26HH105 pKa = 6.62EE106 pKa = 3.97PHH108 pKa = 6.54PHH110 pKa = 5.61PHH112 pKa = 6.57RR113 pKa = 11.84RR114 pKa = 11.84PHH116 pKa = 5.1QQNQQEE122 pKa = 4.6HH123 pKa = 6.06RR124 pKa = 11.84RR125 pKa = 11.84HH126 pKa = 5.99HH127 pKa = 5.6RR128 pKa = 11.84TQHH131 pKa = 4.87RR132 pKa = 11.84RR133 pKa = 11.84RR134 pKa = 11.84PRR136 pKa = 11.84LPLLTRR142 pKa = 11.84HH143 pKa = 5.83RR144 pKa = 11.84HH145 pKa = 4.95RR146 pKa = 11.84PHH148 pKa = 6.44PRR150 pKa = 11.84HH151 pKa = 5.23RR152 pKa = 11.84TPPRR156 pKa = 11.84RR157 pKa = 11.84RR158 pKa = 11.84RR159 pKa = 11.84LRR161 pKa = 11.84RR162 pKa = 11.84TRR164 pKa = 11.84PVRR167 pKa = 11.84RR168 pKa = 11.84IPRR171 pKa = 11.84PPHH174 pKa = 5.63PRR176 pKa = 11.84TRR178 pKa = 11.84PRR180 pKa = 11.84ARR182 pKa = 11.84QRR184 pKa = 11.84HH185 pKa = 6.26RR186 pKa = 11.84IQQPQRR192 pKa = 11.84PLTTRR197 pKa = 11.84RR198 pKa = 11.84RR199 pKa = 11.84PHH201 pKa = 5.38HH202 pKa = 4.47QTRR205 pKa = 11.84RR206 pKa = 11.84RR207 pKa = 11.84PRR209 pKa = 11.84RR210 pKa = 11.84TRR212 pKa = 11.84RR213 pKa = 11.84HH214 pKa = 5.06IRR216 pKa = 11.84GHH218 pKa = 4.61HH219 pKa = 6.29HH220 pKa = 7.44RR221 pKa = 11.84LTADD225 pKa = 3.63PLAPTPLHH233 pKa = 6.36RR234 pKa = 11.84RR235 pKa = 11.84SEE237 pKa = 4.22PVPRR241 pKa = 11.84RR242 pKa = 11.84GIARR246 pKa = 11.84RR247 pKa = 11.84TPPQQRR253 pKa = 11.84HH254 pKa = 4.54RR255 pKa = 11.84HH256 pKa = 4.48PTPRR260 pKa = 11.84GIHH263 pKa = 5.86PRR265 pKa = 11.84VHH267 pKa = 5.47HH268 pKa = 5.83TRR270 pKa = 11.84RR271 pKa = 11.84TLPQPAPQPVPTQLRR286 pKa = 11.84RR287 pKa = 11.84ITRR290 pKa = 11.84PQRR293 pKa = 11.84PTHH296 pKa = 5.15PHH298 pKa = 5.78PRR300 pKa = 11.84HH301 pKa = 6.05PARR304 pKa = 11.84PSAPPPTTRR313 pKa = 11.84RR314 pKa = 11.84TVVSQQRR321 pKa = 11.84HH322 pKa = 3.94RR323 pKa = 11.84QNRR326 pKa = 11.84ARR328 pKa = 11.84RR329 pKa = 11.84PRR331 pKa = 11.84AAAPTTGTDD340 pKa = 2.84VMRR343 pKa = 11.84RR344 pKa = 11.84GDD346 pKa = 3.42SS347 pKa = 3.16
MM1 pKa = 7.34NPAIHH6 pKa = 6.77RR7 pKa = 11.84RR8 pKa = 11.84HH9 pKa = 5.56TPALIPNSAPTPPKK23 pKa = 7.0TTPRR27 pKa = 11.84ASQQVAGIHH36 pKa = 5.21RR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84TRR42 pKa = 11.84PRR44 pKa = 11.84HH45 pKa = 4.87HH46 pKa = 6.97LTRR49 pKa = 11.84IPRR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84LHH56 pKa = 5.53PTRR59 pKa = 11.84LTRR62 pKa = 11.84PRR64 pKa = 11.84RR65 pKa = 11.84RR66 pKa = 11.84RR67 pKa = 11.84RR68 pKa = 11.84PRR70 pKa = 11.84LPTTPRR76 pKa = 11.84RR77 pKa = 11.84TRR79 pKa = 11.84RR80 pKa = 11.84PPTRR84 pKa = 11.84RR85 pKa = 11.84RR86 pKa = 11.84RR87 pKa = 11.84RR88 pKa = 11.84RR89 pKa = 11.84RR90 pKa = 11.84RR91 pKa = 11.84NRR93 pKa = 11.84RR94 pKa = 11.84RR95 pKa = 11.84RR96 pKa = 11.84RR97 pKa = 11.84HH98 pKa = 4.61LRR100 pKa = 11.84LRR102 pKa = 11.84RR103 pKa = 11.84HH104 pKa = 6.26HH105 pKa = 6.62EE106 pKa = 3.97PHH108 pKa = 6.54PHH110 pKa = 5.61PHH112 pKa = 6.57RR113 pKa = 11.84RR114 pKa = 11.84PHH116 pKa = 5.1QQNQQEE122 pKa = 4.6HH123 pKa = 6.06RR124 pKa = 11.84RR125 pKa = 11.84HH126 pKa = 5.99HH127 pKa = 5.6RR128 pKa = 11.84TQHH131 pKa = 4.87RR132 pKa = 11.84RR133 pKa = 11.84RR134 pKa = 11.84PRR136 pKa = 11.84LPLLTRR142 pKa = 11.84HH143 pKa = 5.83RR144 pKa = 11.84HH145 pKa = 4.95RR146 pKa = 11.84PHH148 pKa = 6.44PRR150 pKa = 11.84HH151 pKa = 5.23RR152 pKa = 11.84TPPRR156 pKa = 11.84RR157 pKa = 11.84RR158 pKa = 11.84RR159 pKa = 11.84LRR161 pKa = 11.84RR162 pKa = 11.84TRR164 pKa = 11.84PVRR167 pKa = 11.84RR168 pKa = 11.84IPRR171 pKa = 11.84PPHH174 pKa = 5.63PRR176 pKa = 11.84TRR178 pKa = 11.84PRR180 pKa = 11.84ARR182 pKa = 11.84QRR184 pKa = 11.84HH185 pKa = 6.26RR186 pKa = 11.84IQQPQRR192 pKa = 11.84PLTTRR197 pKa = 11.84RR198 pKa = 11.84RR199 pKa = 11.84PHH201 pKa = 5.38HH202 pKa = 4.47QTRR205 pKa = 11.84RR206 pKa = 11.84RR207 pKa = 11.84PRR209 pKa = 11.84RR210 pKa = 11.84TRR212 pKa = 11.84RR213 pKa = 11.84HH214 pKa = 5.06IRR216 pKa = 11.84GHH218 pKa = 4.61HH219 pKa = 6.29HH220 pKa = 7.44RR221 pKa = 11.84LTADD225 pKa = 3.63PLAPTPLHH233 pKa = 6.36RR234 pKa = 11.84RR235 pKa = 11.84SEE237 pKa = 4.22PVPRR241 pKa = 11.84RR242 pKa = 11.84GIARR246 pKa = 11.84RR247 pKa = 11.84TPPQQRR253 pKa = 11.84HH254 pKa = 4.54RR255 pKa = 11.84HH256 pKa = 4.48PTPRR260 pKa = 11.84GIHH263 pKa = 5.86PRR265 pKa = 11.84VHH267 pKa = 5.47HH268 pKa = 5.83TRR270 pKa = 11.84RR271 pKa = 11.84TLPQPAPQPVPTQLRR286 pKa = 11.84RR287 pKa = 11.84ITRR290 pKa = 11.84PQRR293 pKa = 11.84PTHH296 pKa = 5.15PHH298 pKa = 5.78PRR300 pKa = 11.84HH301 pKa = 6.05PARR304 pKa = 11.84PSAPPPTTRR313 pKa = 11.84RR314 pKa = 11.84TVVSQQRR321 pKa = 11.84HH322 pKa = 3.94RR323 pKa = 11.84QNRR326 pKa = 11.84ARR328 pKa = 11.84RR329 pKa = 11.84PRR331 pKa = 11.84AAAPTTGTDD340 pKa = 2.84VMRR343 pKa = 11.84RR344 pKa = 11.84GDD346 pKa = 3.42SS347 pKa = 3.16
Molecular weight: 42.76 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2018764 |
29 |
7340 |
320.4 |
34.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.891 ± 0.058 | 0.719 ± 0.008 |
5.954 ± 0.023 | 5.98 ± 0.03 |
2.676 ± 0.016 | 9.768 ± 0.033 |
2.143 ± 0.014 | 3.078 ± 0.02 |
1.207 ± 0.017 | 10.494 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.665 ± 0.013 | 1.541 ± 0.017 |
6.551 ± 0.029 | 2.429 ± 0.018 |
8.84 ± 0.041 | 4.63 ± 0.02 |
5.396 ± 0.024 | 8.51 ± 0.031 |
1.526 ± 0.013 | 2.001 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
