
Pacific flying fox faeces associated circular DNA virus-7
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 8.13
Get precalculated fractions of proteins
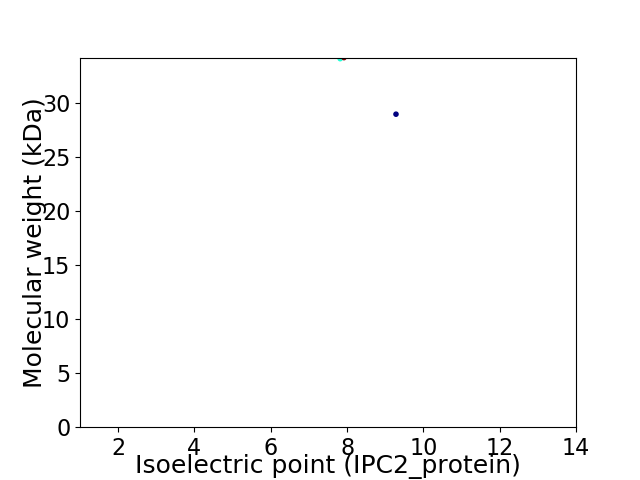
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140CTU3|A0A140CTU3_9VIRU Putative capsid protein OS=Pacific flying fox faeces associated circular DNA virus-7 OX=1796016 PE=4 SV=1
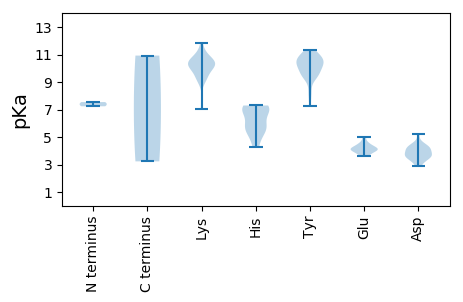
MM1 pKa = 7.24GADD4 pKa = 4.07GNHH7 pKa = 6.95HH8 pKa = 4.88YY9 pKa = 10.22QGYY12 pKa = 9.38IEE14 pKa = 4.61MPRR17 pKa = 11.84SVKK20 pKa = 8.9FTHH23 pKa = 6.5FKK25 pKa = 10.53CLAGAHH31 pKa = 5.68FEE33 pKa = 4.44KK34 pKa = 10.56RR35 pKa = 11.84WGTAQQASDD44 pKa = 3.73YY45 pKa = 11.32CNGLDD50 pKa = 4.28HH51 pKa = 7.29KK52 pKa = 11.25KK53 pKa = 10.4PGDD56 pKa = 3.47KK57 pKa = 10.64SIVIDD62 pKa = 5.25GPWIWGTMSKK72 pKa = 10.7GQGSRR77 pKa = 11.84TDD79 pKa = 3.38VLNLRR84 pKa = 11.84DD85 pKa = 3.59AVKK88 pKa = 9.99GGKK91 pKa = 9.25RR92 pKa = 11.84GRR94 pKa = 11.84EE95 pKa = 4.06LYY97 pKa = 10.83DD98 pKa = 4.48DD99 pKa = 4.17DD100 pKa = 4.58TVAGAAIKK108 pKa = 10.22FSRR111 pKa = 11.84GVAEE115 pKa = 4.05LTRR118 pKa = 11.84AYY120 pKa = 9.35STAPSRR126 pKa = 11.84GDD128 pKa = 3.03IRR130 pKa = 11.84CIFHH134 pKa = 7.03FGPPGTGKK142 pKa = 7.05THH144 pKa = 7.08CAHH147 pKa = 7.29DD148 pKa = 4.2DD149 pKa = 3.38KK150 pKa = 11.82GIFNINLTITLTLYY164 pKa = 9.37IAYY167 pKa = 10.19YY168 pKa = 10.6FDD170 pKa = 4.09GNNGGFWIGYY180 pKa = 8.05QGEE183 pKa = 4.17EE184 pKa = 3.88TAILDD189 pKa = 3.67EE190 pKa = 4.99FGGHH194 pKa = 5.6TLPPKK199 pKa = 9.99QLQRR203 pKa = 11.84LCDD206 pKa = 3.81KK207 pKa = 10.45YY208 pKa = 10.62PYY210 pKa = 9.48WMSIKK215 pKa = 10.24GGEE218 pKa = 4.43VACHH222 pKa = 5.79IRR224 pKa = 11.84TVHH227 pKa = 5.57ICSNYY232 pKa = 9.86LPSGWWSEE240 pKa = 3.67KK241 pKa = 9.38TLYY244 pKa = 11.0NEE246 pKa = 3.66DD247 pKa = 3.6AIYY250 pKa = 10.5RR251 pKa = 11.84RR252 pKa = 11.84IVEE255 pKa = 4.13VHH257 pKa = 4.26WHH259 pKa = 5.69YY260 pKa = 11.12EE261 pKa = 3.92FKK263 pKa = 10.69KK264 pKa = 9.45YY265 pKa = 10.51RR266 pKa = 11.84LYY268 pKa = 10.13KK269 pKa = 10.17TDD271 pKa = 4.4DD272 pKa = 4.3PLDD275 pKa = 3.82KK276 pKa = 10.57STWAMYY282 pKa = 10.63KK283 pKa = 9.98FLKK286 pKa = 10.64AKK288 pKa = 10.66LEE290 pKa = 4.08LEE292 pKa = 4.28YY293 pKa = 11.0TPIVHH298 pKa = 7.28KK299 pKa = 10.92
MM1 pKa = 7.24GADD4 pKa = 4.07GNHH7 pKa = 6.95HH8 pKa = 4.88YY9 pKa = 10.22QGYY12 pKa = 9.38IEE14 pKa = 4.61MPRR17 pKa = 11.84SVKK20 pKa = 8.9FTHH23 pKa = 6.5FKK25 pKa = 10.53CLAGAHH31 pKa = 5.68FEE33 pKa = 4.44KK34 pKa = 10.56RR35 pKa = 11.84WGTAQQASDD44 pKa = 3.73YY45 pKa = 11.32CNGLDD50 pKa = 4.28HH51 pKa = 7.29KK52 pKa = 11.25KK53 pKa = 10.4PGDD56 pKa = 3.47KK57 pKa = 10.64SIVIDD62 pKa = 5.25GPWIWGTMSKK72 pKa = 10.7GQGSRR77 pKa = 11.84TDD79 pKa = 3.38VLNLRR84 pKa = 11.84DD85 pKa = 3.59AVKK88 pKa = 9.99GGKK91 pKa = 9.25RR92 pKa = 11.84GRR94 pKa = 11.84EE95 pKa = 4.06LYY97 pKa = 10.83DD98 pKa = 4.48DD99 pKa = 4.17DD100 pKa = 4.58TVAGAAIKK108 pKa = 10.22FSRR111 pKa = 11.84GVAEE115 pKa = 4.05LTRR118 pKa = 11.84AYY120 pKa = 9.35STAPSRR126 pKa = 11.84GDD128 pKa = 3.03IRR130 pKa = 11.84CIFHH134 pKa = 7.03FGPPGTGKK142 pKa = 7.05THH144 pKa = 7.08CAHH147 pKa = 7.29DD148 pKa = 4.2DD149 pKa = 3.38KK150 pKa = 11.82GIFNINLTITLTLYY164 pKa = 9.37IAYY167 pKa = 10.19YY168 pKa = 10.6FDD170 pKa = 4.09GNNGGFWIGYY180 pKa = 8.05QGEE183 pKa = 4.17EE184 pKa = 3.88TAILDD189 pKa = 3.67EE190 pKa = 4.99FGGHH194 pKa = 5.6TLPPKK199 pKa = 9.99QLQRR203 pKa = 11.84LCDD206 pKa = 3.81KK207 pKa = 10.45YY208 pKa = 10.62PYY210 pKa = 9.48WMSIKK215 pKa = 10.24GGEE218 pKa = 4.43VACHH222 pKa = 5.79IRR224 pKa = 11.84TVHH227 pKa = 5.57ICSNYY232 pKa = 9.86LPSGWWSEE240 pKa = 3.67KK241 pKa = 9.38TLYY244 pKa = 11.0NEE246 pKa = 3.66DD247 pKa = 3.6AIYY250 pKa = 10.5RR251 pKa = 11.84RR252 pKa = 11.84IVEE255 pKa = 4.13VHH257 pKa = 4.26WHH259 pKa = 5.69YY260 pKa = 11.12EE261 pKa = 3.92FKK263 pKa = 10.69KK264 pKa = 9.45YY265 pKa = 10.51RR266 pKa = 11.84LYY268 pKa = 10.13KK269 pKa = 10.17TDD271 pKa = 4.4DD272 pKa = 4.3PLDD275 pKa = 3.82KK276 pKa = 10.57STWAMYY282 pKa = 10.63KK283 pKa = 9.98FLKK286 pKa = 10.64AKK288 pKa = 10.66LEE290 pKa = 4.08LEE292 pKa = 4.28YY293 pKa = 11.0TPIVHH298 pKa = 7.28KK299 pKa = 10.92
Molecular weight: 34.15 kDa
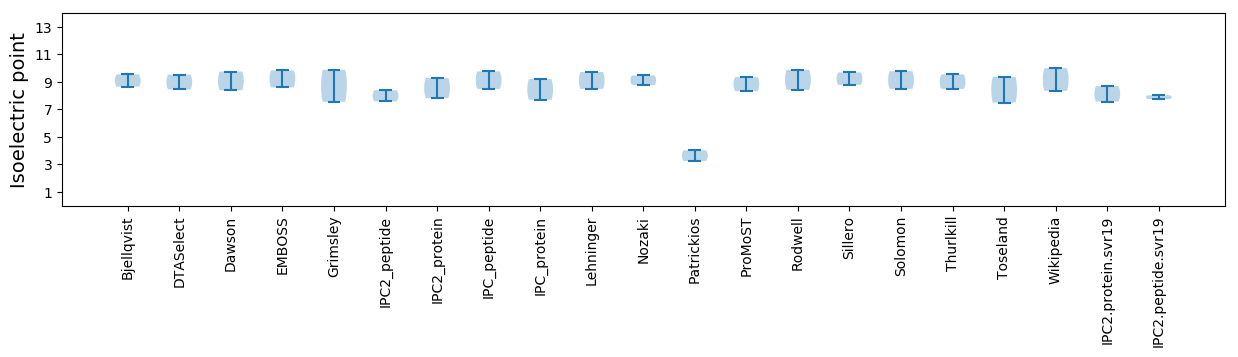
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140CTU3|A0A140CTU3_9VIRU Putative capsid protein OS=Pacific flying fox faeces associated circular DNA virus-7 OX=1796016 PE=4 SV=1
MM1 pKa = 7.54NGLQVRR7 pKa = 11.84QVGIFQSYY15 pKa = 10.57SFSQTPSGTVSVLPSSDD32 pKa = 3.27ANAVPIIIQNQSPSQLSSFINSIAATTDD60 pKa = 2.88TKK62 pKa = 11.18VYY64 pKa = 10.0IRR66 pKa = 11.84QVYY69 pKa = 8.41MKK71 pKa = 10.28GVYY74 pKa = 9.75YY75 pKa = 10.84NNSNTPIIMYY85 pKa = 9.53RR86 pKa = 11.84YY87 pKa = 9.42SFRR90 pKa = 11.84FRR92 pKa = 11.84KK93 pKa = 9.48NVATADD99 pKa = 3.77FASINAILTRR109 pKa = 11.84DD110 pKa = 3.62NPNINDD116 pKa = 4.04PLVNPFTSQIAQRR129 pKa = 11.84LMKK132 pKa = 10.42FGRR135 pKa = 11.84VKK137 pKa = 10.28AQYY140 pKa = 7.24MACGAMGTFKK150 pKa = 11.12LNNKK154 pKa = 9.1YY155 pKa = 10.82YY156 pKa = 10.47NPKK159 pKa = 10.1LVNQEE164 pKa = 4.16TEE166 pKa = 4.14GDD168 pKa = 4.15SVNYY172 pKa = 10.22LAMAGNKK179 pKa = 9.86CYY181 pKa = 9.8MFKK184 pKa = 9.9CTPVPIAHH192 pKa = 6.97FAGATTPVTILGTSWGSWSVSFNYY216 pKa = 9.49MSYY219 pKa = 10.48VSGYY223 pKa = 11.03VIGQNNPVTTYY234 pKa = 10.2VPPFIPTQTNKK245 pKa = 10.12YY246 pKa = 10.22FIFSDD251 pKa = 3.57QTGKK255 pKa = 9.33PQSTTNN261 pKa = 3.23
MM1 pKa = 7.54NGLQVRR7 pKa = 11.84QVGIFQSYY15 pKa = 10.57SFSQTPSGTVSVLPSSDD32 pKa = 3.27ANAVPIIIQNQSPSQLSSFINSIAATTDD60 pKa = 2.88TKK62 pKa = 11.18VYY64 pKa = 10.0IRR66 pKa = 11.84QVYY69 pKa = 8.41MKK71 pKa = 10.28GVYY74 pKa = 9.75YY75 pKa = 10.84NNSNTPIIMYY85 pKa = 9.53RR86 pKa = 11.84YY87 pKa = 9.42SFRR90 pKa = 11.84FRR92 pKa = 11.84KK93 pKa = 9.48NVATADD99 pKa = 3.77FASINAILTRR109 pKa = 11.84DD110 pKa = 3.62NPNINDD116 pKa = 4.04PLVNPFTSQIAQRR129 pKa = 11.84LMKK132 pKa = 10.42FGRR135 pKa = 11.84VKK137 pKa = 10.28AQYY140 pKa = 7.24MACGAMGTFKK150 pKa = 11.12LNNKK154 pKa = 9.1YY155 pKa = 10.82YY156 pKa = 10.47NPKK159 pKa = 10.1LVNQEE164 pKa = 4.16TEE166 pKa = 4.14GDD168 pKa = 4.15SVNYY172 pKa = 10.22LAMAGNKK179 pKa = 9.86CYY181 pKa = 9.8MFKK184 pKa = 9.9CTPVPIAHH192 pKa = 6.97FAGATTPVTILGTSWGSWSVSFNYY216 pKa = 9.49MSYY219 pKa = 10.48VSGYY223 pKa = 11.03VIGQNNPVTTYY234 pKa = 10.2VPPFIPTQTNKK245 pKa = 10.12YY246 pKa = 10.22FIFSDD251 pKa = 3.57QTGKK255 pKa = 9.33PQSTTNN261 pKa = 3.23
Molecular weight: 29.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
560 |
261 |
299 |
280.0 |
31.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.429 ± 0.057 | 1.786 ± 0.428 |
4.821 ± 1.44 | 2.857 ± 1.408 |
4.821 ± 0.623 | 8.214 ± 1.661 |
2.679 ± 1.546 | 6.607 ± 0.195 |
6.429 ± 1.233 | 5.357 ± 1.027 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.5 ± 0.638 | 5.893 ± 2.224 |
5.179 ± 0.899 | 4.107 ± 1.362 |
4.107 ± 0.702 | 6.786 ± 1.88 |
7.5 ± 0.884 | 5.536 ± 1.432 |
1.964 ± 0.807 | 6.429 ± 0.057 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
