
Leptospira biflexa serovar Patoc (strain Patoc 1 / ATCC 23582 / Paris)
Taxonomy: cellular organisms; Bacteria; Spirochaetes; Spirochaetia; Leptospirales; Leptospiraceae; Leptospira; Leptospira biflexa; Leptospira biflexa serovar Patoc
Average proteome isoelectric point is 6.8
Get precalculated fractions of proteins
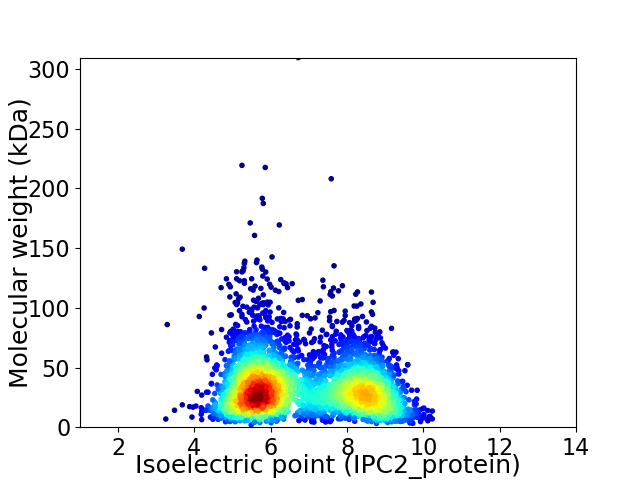
Virtual 2D-PAGE plot for 3723 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B0SP40|B0SP40_LEPBP Uncharacterized protein OS=Leptospira biflexa serovar Patoc (strain Patoc 1 / ATCC 23582 / Paris) OX=456481 GN=LEPBI_I2984 PE=4 SV=1
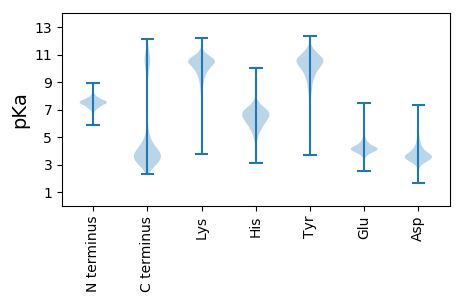
MM1 pKa = 7.39GFIRR5 pKa = 11.84GQFGFVFKK13 pKa = 11.13LKK15 pKa = 8.8FTHH18 pKa = 6.44VLFLFTTLFVSCQSVTPVNLQMLFGSFFVSATGQGSVIYY57 pKa = 7.85EE58 pKa = 3.89APNFLFTSEE67 pKa = 4.16NGKK70 pKa = 8.72QAEE73 pKa = 4.26FNLRR77 pKa = 11.84LNMEE81 pKa = 4.71PNSSVRR87 pKa = 11.84IGPITISDD95 pKa = 3.64PTEE98 pKa = 3.78AVLVSDD104 pKa = 3.59TFINFTEE111 pKa = 4.2EE112 pKa = 4.07DD113 pKa = 3.54WNEE116 pKa = 3.6PHH118 pKa = 7.25IVRR121 pKa = 11.84LVGIDD126 pKa = 4.38DD127 pKa = 4.44PFSDD131 pKa = 4.0GNQNYY136 pKa = 8.94RR137 pKa = 11.84VQLGSILTSDD147 pKa = 3.51IRR149 pKa = 11.84FSTQSLPVLLVVNTDD164 pKa = 3.49NEE166 pKa = 4.6SSGVAASPVFGLLTSEE182 pKa = 4.18TGEE185 pKa = 4.24TGQISYY191 pKa = 10.73VLQTRR196 pKa = 11.84PMQDD200 pKa = 2.35VYY202 pKa = 11.5LRR204 pKa = 11.84NFVSSDD210 pKa = 3.05TTEE213 pKa = 3.89ATVEE217 pKa = 4.16SVEE220 pKa = 5.72LVFTPNNWDD229 pKa = 3.32VPQSVTVTGVDD240 pKa = 3.75DD241 pKa = 4.71FSVDD245 pKa = 3.03NSNFQISADD254 pKa = 3.59ATVSMDD260 pKa = 3.45PAYY263 pKa = 10.39LGKK266 pKa = 10.19VVPIITGTNVDD277 pKa = 3.17NDD279 pKa = 3.37IAGFTVVNLSGLTTTEE295 pKa = 3.49AGGAISFGVVLNTLPTHH312 pKa = 6.23SVTIPSIVATPTTEE326 pKa = 4.18SSISPTTLTFAPGEE340 pKa = 4.13WFTPKK345 pKa = 9.86IVTVTGVDD353 pKa = 3.45EE354 pKa = 5.46FIVDD358 pKa = 4.1GSQTVTIVSSAATSTDD374 pKa = 2.43SDD376 pKa = 4.67YY377 pKa = 11.81NGLAGPVFPSVTNTDD392 pKa = 2.77NDD394 pKa = 3.52VPGFVLTPPGSLSISEE410 pKa = 4.21NGGVLNFNIHH420 pKa = 6.91LSSQPPPGFTVTLTGINEE438 pKa = 4.18NNSITNVNTSSLVFTNANWNLDD460 pKa = 3.26QTIQITTNNNDD471 pKa = 2.71VDD473 pKa = 4.01EE474 pKa = 4.6DD475 pKa = 4.13TRR477 pKa = 11.84TVTLQFGSVDD487 pKa = 3.5TGGSADD493 pKa = 3.63PVYY496 pKa = 10.97NAIPPPTQVSFSVTDD511 pKa = 4.26DD512 pKa = 3.37DD513 pKa = 4.36TAGITVTPVGGLVVHH528 pKa = 6.67EE529 pKa = 4.8NGTPFTEE536 pKa = 4.4TFTVVLNSQPTQTVNIPTISSSNTAEE562 pKa = 3.98ITVSPSSLSFTTANWNTPQTVTLTSVLDD590 pKa = 4.21GSDD593 pKa = 5.68DD594 pKa = 3.48GDD596 pKa = 3.61QNVNINLSNTSSTDD610 pKa = 3.05PKK612 pKa = 10.76YY613 pKa = 11.01NSIVIASVTAINTDD627 pKa = 3.28SNEE630 pKa = 3.84PLVRR634 pKa = 11.84IQNLSASSIVEE645 pKa = 4.57DD646 pKa = 3.88GTSTITFEE654 pKa = 4.43IRR656 pKa = 11.84LSLKK660 pKa = 10.09PNANVTIGPITSSDD674 pKa = 3.07GTEE677 pKa = 3.53AVLLNSTSGVAASRR691 pKa = 11.84TLTFTPTNAQVANYY705 pKa = 9.81SGNTSEE711 pKa = 5.4SGWDD715 pKa = 3.52VPQTITIRR723 pKa = 11.84SVADD727 pKa = 3.46SFDD730 pKa = 4.14DD731 pKa = 4.17GNLPVTIHH739 pKa = 6.72IPQASGSYY747 pKa = 7.68FTGLYY752 pKa = 6.68PTGAVPGYY760 pKa = 9.43TDD762 pKa = 3.19TNGNLVVTVTDD773 pKa = 3.53NDD775 pKa = 4.07TKK777 pKa = 11.33GFTFSTTTLNLTEE790 pKa = 4.65GDD792 pKa = 3.19VDD794 pKa = 3.67GTFTVRR800 pKa = 11.84LNAAPCDD807 pKa = 4.06TPGNLAVCASGSVTIPISAEE827 pKa = 4.11TFSLPDD833 pKa = 3.26SAQYY837 pKa = 9.22TVSPANLTFTHH848 pKa = 6.49TNFVTPQTVTVTVVNDD864 pKa = 4.43SINEE868 pKa = 4.37SNTRR872 pKa = 11.84THH874 pKa = 6.46TLTLGAISGSGTDD887 pKa = 3.71YY888 pKa = 11.67EE889 pKa = 4.85GMNPSDD895 pKa = 3.54VTINITDD902 pKa = 3.74NDD904 pKa = 3.92NPSPKK909 pKa = 9.61ILFTLDD915 pKa = 2.58SGQPYY920 pKa = 7.75FTTEE924 pKa = 3.72SGFSTFYY931 pKa = 10.94SLRR934 pKa = 11.84LGSRR938 pKa = 11.84PIPGNSVTVTLSTSDD953 pKa = 3.3TTEE956 pKa = 3.76GMINDD961 pKa = 4.12SGTPASSKK969 pKa = 10.72QYY971 pKa = 10.83IFDD974 pKa = 3.51EE975 pKa = 5.05TNWSTSIPVEE985 pKa = 3.8IVGVLDD991 pKa = 3.68ILSDD995 pKa = 3.75GNINFNITVSGAEE1008 pKa = 4.11TGSFPSWYY1016 pKa = 9.99DD1017 pKa = 3.12SFVGSNGTTANLVNYY1032 pKa = 9.26SVSEE1036 pKa = 4.11NPVTVVTPQSMVRR1049 pKa = 11.84AEE1051 pKa = 3.95NAVAFSIYY1059 pKa = 10.06ILLSQAPTDD1068 pKa = 4.11DD1069 pKa = 3.32VTIPISISSSFPCTLFTGPTVSQFTLSTNTITLTSANWNTVGAHH1113 pKa = 4.56NTITVTPFDD1122 pKa = 5.13DD1123 pKa = 4.85SVDD1126 pKa = 4.07DD1127 pKa = 4.87GNVTCPIVVGNVSSSDD1143 pKa = 3.27GFFNGVNPFPSANYY1157 pKa = 9.8PEE1159 pKa = 4.78LTLNDD1164 pKa = 3.43NDD1166 pKa = 4.06AAGITTSGFSPSTVITSQSGASSEE1190 pKa = 4.61FYY1192 pKa = 10.37IHH1194 pKa = 7.13LNSQPTSDD1202 pKa = 3.43VTINLSAAPGGLVSFPTAPLTFTPSNFGTGQLVTILGQNTADD1244 pKa = 4.16LSDD1247 pKa = 3.53VNYY1250 pKa = 10.77NITPQITSSEE1260 pKa = 4.34SGTGFSPSTIYY1271 pKa = 10.65SALTPSTIPGVHH1283 pKa = 6.96IYY1285 pKa = 10.65NLYY1288 pKa = 10.69DD1289 pKa = 4.78IIPCTDD1295 pKa = 4.1PNPMVACGTSPNGSGGLVTSPNLVTSEE1322 pKa = 4.92LGAQSRR1328 pKa = 11.84FQLRR1332 pKa = 11.84FRR1334 pKa = 11.84ARR1336 pKa = 11.84PTSNVTIGVASSNASEE1352 pKa = 4.46GTTAVPNVTFTPTNWNTFQNIVITGVDD1379 pKa = 3.56DD1380 pKa = 4.22GLVDD1384 pKa = 4.23GNVLYY1389 pKa = 11.0SILFGSLTGGGTGFNGEE1406 pKa = 4.4SLPNVTVTNQDD1417 pKa = 2.5NDD1419 pKa = 3.38
MM1 pKa = 7.39GFIRR5 pKa = 11.84GQFGFVFKK13 pKa = 11.13LKK15 pKa = 8.8FTHH18 pKa = 6.44VLFLFTTLFVSCQSVTPVNLQMLFGSFFVSATGQGSVIYY57 pKa = 7.85EE58 pKa = 3.89APNFLFTSEE67 pKa = 4.16NGKK70 pKa = 8.72QAEE73 pKa = 4.26FNLRR77 pKa = 11.84LNMEE81 pKa = 4.71PNSSVRR87 pKa = 11.84IGPITISDD95 pKa = 3.64PTEE98 pKa = 3.78AVLVSDD104 pKa = 3.59TFINFTEE111 pKa = 4.2EE112 pKa = 4.07DD113 pKa = 3.54WNEE116 pKa = 3.6PHH118 pKa = 7.25IVRR121 pKa = 11.84LVGIDD126 pKa = 4.38DD127 pKa = 4.44PFSDD131 pKa = 4.0GNQNYY136 pKa = 8.94RR137 pKa = 11.84VQLGSILTSDD147 pKa = 3.51IRR149 pKa = 11.84FSTQSLPVLLVVNTDD164 pKa = 3.49NEE166 pKa = 4.6SSGVAASPVFGLLTSEE182 pKa = 4.18TGEE185 pKa = 4.24TGQISYY191 pKa = 10.73VLQTRR196 pKa = 11.84PMQDD200 pKa = 2.35VYY202 pKa = 11.5LRR204 pKa = 11.84NFVSSDD210 pKa = 3.05TTEE213 pKa = 3.89ATVEE217 pKa = 4.16SVEE220 pKa = 5.72LVFTPNNWDD229 pKa = 3.32VPQSVTVTGVDD240 pKa = 3.75DD241 pKa = 4.71FSVDD245 pKa = 3.03NSNFQISADD254 pKa = 3.59ATVSMDD260 pKa = 3.45PAYY263 pKa = 10.39LGKK266 pKa = 10.19VVPIITGTNVDD277 pKa = 3.17NDD279 pKa = 3.37IAGFTVVNLSGLTTTEE295 pKa = 3.49AGGAISFGVVLNTLPTHH312 pKa = 6.23SVTIPSIVATPTTEE326 pKa = 4.18SSISPTTLTFAPGEE340 pKa = 4.13WFTPKK345 pKa = 9.86IVTVTGVDD353 pKa = 3.45EE354 pKa = 5.46FIVDD358 pKa = 4.1GSQTVTIVSSAATSTDD374 pKa = 2.43SDD376 pKa = 4.67YY377 pKa = 11.81NGLAGPVFPSVTNTDD392 pKa = 2.77NDD394 pKa = 3.52VPGFVLTPPGSLSISEE410 pKa = 4.21NGGVLNFNIHH420 pKa = 6.91LSSQPPPGFTVTLTGINEE438 pKa = 4.18NNSITNVNTSSLVFTNANWNLDD460 pKa = 3.26QTIQITTNNNDD471 pKa = 2.71VDD473 pKa = 4.01EE474 pKa = 4.6DD475 pKa = 4.13TRR477 pKa = 11.84TVTLQFGSVDD487 pKa = 3.5TGGSADD493 pKa = 3.63PVYY496 pKa = 10.97NAIPPPTQVSFSVTDD511 pKa = 4.26DD512 pKa = 3.37DD513 pKa = 4.36TAGITVTPVGGLVVHH528 pKa = 6.67EE529 pKa = 4.8NGTPFTEE536 pKa = 4.4TFTVVLNSQPTQTVNIPTISSSNTAEE562 pKa = 3.98ITVSPSSLSFTTANWNTPQTVTLTSVLDD590 pKa = 4.21GSDD593 pKa = 5.68DD594 pKa = 3.48GDD596 pKa = 3.61QNVNINLSNTSSTDD610 pKa = 3.05PKK612 pKa = 10.76YY613 pKa = 11.01NSIVIASVTAINTDD627 pKa = 3.28SNEE630 pKa = 3.84PLVRR634 pKa = 11.84IQNLSASSIVEE645 pKa = 4.57DD646 pKa = 3.88GTSTITFEE654 pKa = 4.43IRR656 pKa = 11.84LSLKK660 pKa = 10.09PNANVTIGPITSSDD674 pKa = 3.07GTEE677 pKa = 3.53AVLLNSTSGVAASRR691 pKa = 11.84TLTFTPTNAQVANYY705 pKa = 9.81SGNTSEE711 pKa = 5.4SGWDD715 pKa = 3.52VPQTITIRR723 pKa = 11.84SVADD727 pKa = 3.46SFDD730 pKa = 4.14DD731 pKa = 4.17GNLPVTIHH739 pKa = 6.72IPQASGSYY747 pKa = 7.68FTGLYY752 pKa = 6.68PTGAVPGYY760 pKa = 9.43TDD762 pKa = 3.19TNGNLVVTVTDD773 pKa = 3.53NDD775 pKa = 4.07TKK777 pKa = 11.33GFTFSTTTLNLTEE790 pKa = 4.65GDD792 pKa = 3.19VDD794 pKa = 3.67GTFTVRR800 pKa = 11.84LNAAPCDD807 pKa = 4.06TPGNLAVCASGSVTIPISAEE827 pKa = 4.11TFSLPDD833 pKa = 3.26SAQYY837 pKa = 9.22TVSPANLTFTHH848 pKa = 6.49TNFVTPQTVTVTVVNDD864 pKa = 4.43SINEE868 pKa = 4.37SNTRR872 pKa = 11.84THH874 pKa = 6.46TLTLGAISGSGTDD887 pKa = 3.71YY888 pKa = 11.67EE889 pKa = 4.85GMNPSDD895 pKa = 3.54VTINITDD902 pKa = 3.74NDD904 pKa = 3.92NPSPKK909 pKa = 9.61ILFTLDD915 pKa = 2.58SGQPYY920 pKa = 7.75FTTEE924 pKa = 3.72SGFSTFYY931 pKa = 10.94SLRR934 pKa = 11.84LGSRR938 pKa = 11.84PIPGNSVTVTLSTSDD953 pKa = 3.3TTEE956 pKa = 3.76GMINDD961 pKa = 4.12SGTPASSKK969 pKa = 10.72QYY971 pKa = 10.83IFDD974 pKa = 3.51EE975 pKa = 5.05TNWSTSIPVEE985 pKa = 3.8IVGVLDD991 pKa = 3.68ILSDD995 pKa = 3.75GNINFNITVSGAEE1008 pKa = 4.11TGSFPSWYY1016 pKa = 9.99DD1017 pKa = 3.12SFVGSNGTTANLVNYY1032 pKa = 9.26SVSEE1036 pKa = 4.11NPVTVVTPQSMVRR1049 pKa = 11.84AEE1051 pKa = 3.95NAVAFSIYY1059 pKa = 10.06ILLSQAPTDD1068 pKa = 4.11DD1069 pKa = 3.32VTIPISISSSFPCTLFTGPTVSQFTLSTNTITLTSANWNTVGAHH1113 pKa = 4.56NTITVTPFDD1122 pKa = 5.13DD1123 pKa = 4.85SVDD1126 pKa = 4.07DD1127 pKa = 4.87GNVTCPIVVGNVSSSDD1143 pKa = 3.27GFFNGVNPFPSANYY1157 pKa = 9.8PEE1159 pKa = 4.78LTLNDD1164 pKa = 3.43NDD1166 pKa = 4.06AAGITTSGFSPSTVITSQSGASSEE1190 pKa = 4.61FYY1192 pKa = 10.37IHH1194 pKa = 7.13LNSQPTSDD1202 pKa = 3.43VTINLSAAPGGLVSFPTAPLTFTPSNFGTGQLVTILGQNTADD1244 pKa = 4.16LSDD1247 pKa = 3.53VNYY1250 pKa = 10.77NITPQITSSEE1260 pKa = 4.34SGTGFSPSTIYY1271 pKa = 10.65SALTPSTIPGVHH1283 pKa = 6.96IYY1285 pKa = 10.65NLYY1288 pKa = 10.69DD1289 pKa = 4.78IIPCTDD1295 pKa = 4.1PNPMVACGTSPNGSGGLVTSPNLVTSEE1322 pKa = 4.92LGAQSRR1328 pKa = 11.84FQLRR1332 pKa = 11.84FRR1334 pKa = 11.84ARR1336 pKa = 11.84PTSNVTIGVASSNASEE1352 pKa = 4.46GTTAVPNVTFTPTNWNTFQNIVITGVDD1379 pKa = 3.56DD1380 pKa = 4.22GLVDD1384 pKa = 4.23GNVLYY1389 pKa = 11.0SILFGSLTGGGTGFNGEE1406 pKa = 4.4SLPNVTVTNQDD1417 pKa = 2.5NDD1419 pKa = 3.38
Molecular weight: 149.15 kDa
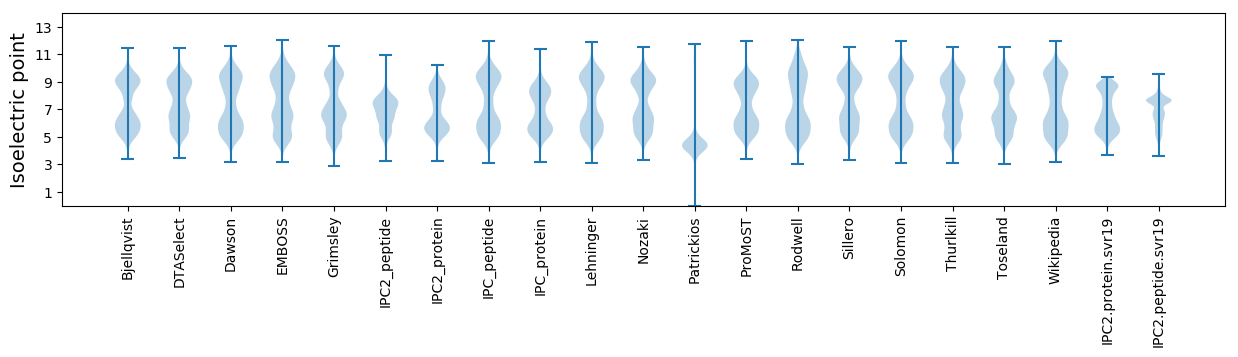
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|B0SJC4|TRUA_LEPBP tRNA pseudouridine synthase A OS=Leptospira biflexa serovar Patoc (strain Patoc 1 / ATCC 23582 / Paris) OX=456481 GN=truA PE=3 SV=1
MM1 pKa = 7.75NSTTKK6 pKa = 10.29KK7 pKa = 9.79DD8 pKa = 3.31YY9 pKa = 10.34WVIGSLLFLSLVPSIAGAVRR29 pKa = 11.84IFQIATGSGYY39 pKa = 7.8TVEE42 pKa = 4.18NQRR45 pKa = 11.84FFNDD49 pKa = 4.94PIPVFVHH56 pKa = 6.39IISVLVYY63 pKa = 10.22SILGAFQFAPGFRR76 pKa = 11.84SRR78 pKa = 11.84HH79 pKa = 4.85LMWHH83 pKa = 5.76RR84 pKa = 11.84VSGRR88 pKa = 11.84FLVLFGLTSAISGVWLTLVYY108 pKa = 10.26PKK110 pKa = 10.76VPTDD114 pKa = 3.67GDD116 pKa = 3.73WLFGIRR122 pKa = 11.84MIVGVWMFLCVSLGFFFVWKK142 pKa = 10.2RR143 pKa = 11.84KK144 pKa = 8.43FQTHH148 pKa = 5.69SNWMLRR154 pKa = 11.84GYY156 pKa = 10.84AIGLGAGTQVFTHH169 pKa = 6.55LPWFVIVGGDD179 pKa = 3.47PSGVPRR185 pKa = 11.84DD186 pKa = 3.79LMMGAGWLINLIFAEE201 pKa = 3.9WLIRR205 pKa = 11.84RR206 pKa = 11.84KK207 pKa = 9.9KK208 pKa = 10.16
MM1 pKa = 7.75NSTTKK6 pKa = 10.29KK7 pKa = 9.79DD8 pKa = 3.31YY9 pKa = 10.34WVIGSLLFLSLVPSIAGAVRR29 pKa = 11.84IFQIATGSGYY39 pKa = 7.8TVEE42 pKa = 4.18NQRR45 pKa = 11.84FFNDD49 pKa = 4.94PIPVFVHH56 pKa = 6.39IISVLVYY63 pKa = 10.22SILGAFQFAPGFRR76 pKa = 11.84SRR78 pKa = 11.84HH79 pKa = 4.85LMWHH83 pKa = 5.76RR84 pKa = 11.84VSGRR88 pKa = 11.84FLVLFGLTSAISGVWLTLVYY108 pKa = 10.26PKK110 pKa = 10.76VPTDD114 pKa = 3.67GDD116 pKa = 3.73WLFGIRR122 pKa = 11.84MIVGVWMFLCVSLGFFFVWKK142 pKa = 10.2RR143 pKa = 11.84KK144 pKa = 8.43FQTHH148 pKa = 5.69SNWMLRR154 pKa = 11.84GYY156 pKa = 10.84AIGLGAGTQVFTHH169 pKa = 6.55LPWFVIVGGDD179 pKa = 3.47PSGVPRR185 pKa = 11.84DD186 pKa = 3.79LMMGAGWLINLIFAEE201 pKa = 3.9WLIRR205 pKa = 11.84RR206 pKa = 11.84KK207 pKa = 9.9KK208 pKa = 10.16
Molecular weight: 23.58 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1222038 |
17 |
2644 |
328.2 |
37.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.492 ± 0.038 | 0.797 ± 0.012 |
4.773 ± 0.025 | 7.052 ± 0.041 |
5.78 ± 0.041 | 6.481 ± 0.036 |
1.907 ± 0.019 | 7.621 ± 0.037 |
7.554 ± 0.043 | 10.419 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.16 ± 0.016 | 4.781 ± 0.031 |
4.075 ± 0.024 | 3.448 ± 0.019 |
4.116 ± 0.024 | 7.38 ± 0.036 |
5.342 ± 0.032 | 6.046 ± 0.032 |
1.135 ± 0.016 | 3.643 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
