
Propionispora vibrioides
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Negativicutes; Selenomonadales; Sporomusaceae; Propionispora
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
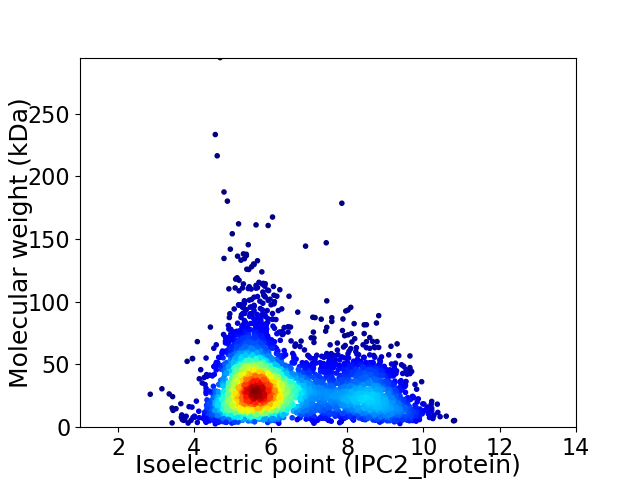
Virtual 2D-PAGE plot for 4126 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H8TK23|A0A1H8TK23_9FIRM Predicted amino acid dehydrogenase OS=Propionispora vibrioides OX=112903 GN=SAMN04490178_1073 PE=4 SV=1
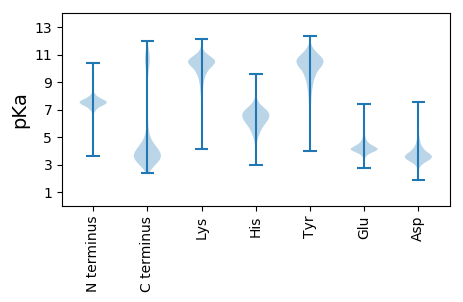
MM1 pKa = 6.96MRR3 pKa = 11.84ALYY6 pKa = 10.45SAVSGIKK13 pKa = 9.87AQQTSLDD20 pKa = 3.94VISNNIANVDD30 pKa = 3.45TTGYY34 pKa = 10.07KK35 pKa = 8.49SQRR38 pKa = 11.84VSFSDD43 pKa = 4.25LLSQTLSSASGSNGTSGGTNPVQVGLGTSVASTDD77 pKa = 3.39TIMTVGSSQSTGVATDD93 pKa = 4.01LSISGNGFFIVTGGSQSQYY112 pKa = 9.85QFTRR116 pKa = 11.84EE117 pKa = 3.47GDD119 pKa = 3.4LTYY122 pKa = 10.99DD123 pKa = 3.4ADD125 pKa = 4.54GNLTINGYY133 pKa = 8.37KK134 pKa = 8.66VCGWEE139 pKa = 4.67SYY141 pKa = 7.63TTDD144 pKa = 3.01SDD146 pKa = 3.94GNKK149 pKa = 9.78VYY151 pKa = 9.3NTNGAVEE158 pKa = 4.87PINLYY163 pKa = 10.45SDD165 pKa = 4.04SYY167 pKa = 11.41SGNKK171 pKa = 9.72KK172 pKa = 10.09LMAAKK177 pKa = 8.84ATTKK181 pKa = 8.74EE182 pKa = 4.32TLSGNLKK189 pKa = 7.7TTATAKK195 pKa = 9.21GTALNNIGTISQTATTKK212 pKa = 10.15TNSTGVSVSSASGLTQNSTYY232 pKa = 10.62TVTLTDD238 pKa = 3.82GTTSGHH244 pKa = 6.41FDD246 pKa = 2.96ITVTDD251 pKa = 3.79ASGATVATLTDD262 pKa = 3.61EE263 pKa = 4.81DD264 pKa = 4.91LSDD267 pKa = 3.78GATLTGSNGEE277 pKa = 4.51SIVLASNSSATAGTLTFTAANSKK300 pKa = 10.54DD301 pKa = 3.29QTTTMTVYY309 pKa = 10.51DD310 pKa = 4.21AQGNSYY316 pKa = 10.5DD317 pKa = 3.54VSVNFTKK324 pKa = 10.14SYY326 pKa = 9.7YY327 pKa = 10.41DD328 pKa = 3.12SSTGQTSWYY337 pKa = 8.67WEE339 pKa = 4.19TSSSDD344 pKa = 3.66SNVSSVTGSGYY355 pKa = 11.16LLFDD359 pKa = 3.34SSGNLVTTDD368 pKa = 2.69SSYY371 pKa = 11.49PSTASVTVSPSGSSPVTLSLDD392 pKa = 3.99FSNVASMVLSSGDD405 pKa = 3.49SSVTSSQDD413 pKa = 2.54GYY415 pKa = 11.82AAGTLSSLSISSDD428 pKa = 2.97GTITGTYY435 pKa = 10.65SNGQTQSLAQIALANFTNPQGLEE458 pKa = 3.96KK459 pKa = 11.01VGDD462 pKa = 3.87NLYY465 pKa = 9.05VTTVNSGAFTGGVVAGSNGTGSLSSGTLEE494 pKa = 3.98MSNVDD499 pKa = 4.06LAEE502 pKa = 3.99QFSNMMISQRR512 pKa = 11.84AYY514 pKa = 8.69QANSKK519 pKa = 10.54VITAADD525 pKa = 3.27QCLQSLINMVSS536 pKa = 2.82
MM1 pKa = 6.96MRR3 pKa = 11.84ALYY6 pKa = 10.45SAVSGIKK13 pKa = 9.87AQQTSLDD20 pKa = 3.94VISNNIANVDD30 pKa = 3.45TTGYY34 pKa = 10.07KK35 pKa = 8.49SQRR38 pKa = 11.84VSFSDD43 pKa = 4.25LLSQTLSSASGSNGTSGGTNPVQVGLGTSVASTDD77 pKa = 3.39TIMTVGSSQSTGVATDD93 pKa = 4.01LSISGNGFFIVTGGSQSQYY112 pKa = 9.85QFTRR116 pKa = 11.84EE117 pKa = 3.47GDD119 pKa = 3.4LTYY122 pKa = 10.99DD123 pKa = 3.4ADD125 pKa = 4.54GNLTINGYY133 pKa = 8.37KK134 pKa = 8.66VCGWEE139 pKa = 4.67SYY141 pKa = 7.63TTDD144 pKa = 3.01SDD146 pKa = 3.94GNKK149 pKa = 9.78VYY151 pKa = 9.3NTNGAVEE158 pKa = 4.87PINLYY163 pKa = 10.45SDD165 pKa = 4.04SYY167 pKa = 11.41SGNKK171 pKa = 9.72KK172 pKa = 10.09LMAAKK177 pKa = 8.84ATTKK181 pKa = 8.74EE182 pKa = 4.32TLSGNLKK189 pKa = 7.7TTATAKK195 pKa = 9.21GTALNNIGTISQTATTKK212 pKa = 10.15TNSTGVSVSSASGLTQNSTYY232 pKa = 10.62TVTLTDD238 pKa = 3.82GTTSGHH244 pKa = 6.41FDD246 pKa = 2.96ITVTDD251 pKa = 3.79ASGATVATLTDD262 pKa = 3.61EE263 pKa = 4.81DD264 pKa = 4.91LSDD267 pKa = 3.78GATLTGSNGEE277 pKa = 4.51SIVLASNSSATAGTLTFTAANSKK300 pKa = 10.54DD301 pKa = 3.29QTTTMTVYY309 pKa = 10.51DD310 pKa = 4.21AQGNSYY316 pKa = 10.5DD317 pKa = 3.54VSVNFTKK324 pKa = 10.14SYY326 pKa = 9.7YY327 pKa = 10.41DD328 pKa = 3.12SSTGQTSWYY337 pKa = 8.67WEE339 pKa = 4.19TSSSDD344 pKa = 3.66SNVSSVTGSGYY355 pKa = 11.16LLFDD359 pKa = 3.34SSGNLVTTDD368 pKa = 2.69SSYY371 pKa = 11.49PSTASVTVSPSGSSPVTLSLDD392 pKa = 3.99FSNVASMVLSSGDD405 pKa = 3.49SSVTSSQDD413 pKa = 2.54GYY415 pKa = 11.82AAGTLSSLSISSDD428 pKa = 2.97GTITGTYY435 pKa = 10.65SNGQTQSLAQIALANFTNPQGLEE458 pKa = 3.96KK459 pKa = 11.01VGDD462 pKa = 3.87NLYY465 pKa = 9.05VTTVNSGAFTGGVVAGSNGTGSLSSGTLEE494 pKa = 3.98MSNVDD499 pKa = 4.06LAEE502 pKa = 3.99QFSNMMISQRR512 pKa = 11.84AYY514 pKa = 8.69QANSKK519 pKa = 10.54VITAADD525 pKa = 3.27QCLQSLINMVSS536 pKa = 2.82
Molecular weight: 54.77 kDa
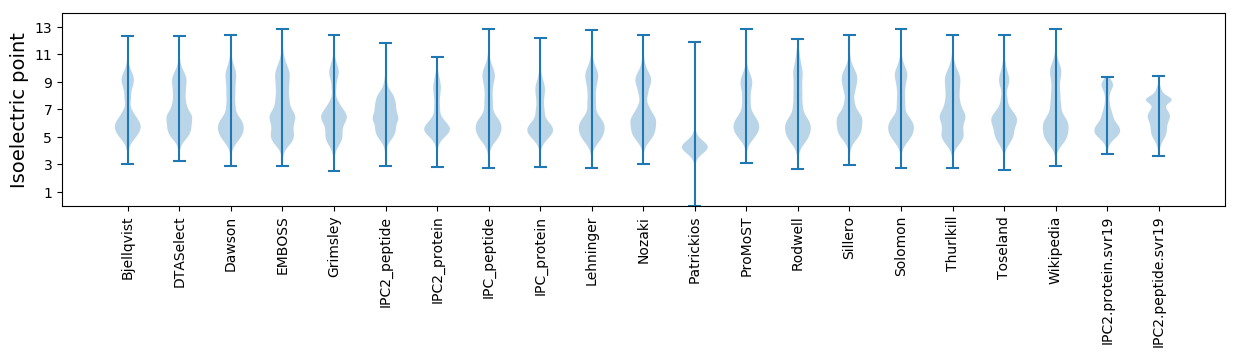
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H8XL13|A0A1H8XL13_9FIRM Aspartate ammonia-lyase OS=Propionispora vibrioides OX=112903 GN=SAMN04490178_12547 PE=4 SV=1
MM1 pKa = 7.75RR2 pKa = 11.84EE3 pKa = 3.65NVRR6 pKa = 11.84WMTEE10 pKa = 3.85VALLAAFLSVAGAFKK25 pKa = 10.99LPGLLPGTEE34 pKa = 4.38FQLSAPLAVAICAVFGFAKK53 pKa = 10.65YY54 pKa = 8.25ITAGLLSSMAGLMLGTQSVWNVGIALIFRR83 pKa = 11.84CVVGAVFVLGGNRR96 pKa = 11.84WLTVVLAGPIATITSRR112 pKa = 11.84LVVGGILGNMAIPFMLAAVPGIIYY136 pKa = 8.09TAITVWPLTILLKK149 pKa = 10.68RR150 pKa = 11.84ITEE153 pKa = 4.1RR154 pKa = 11.84GKK156 pKa = 9.85KK157 pKa = 9.77AIVYY161 pKa = 7.51VVQRR165 pKa = 3.91
MM1 pKa = 7.75RR2 pKa = 11.84EE3 pKa = 3.65NVRR6 pKa = 11.84WMTEE10 pKa = 3.85VALLAAFLSVAGAFKK25 pKa = 10.99LPGLLPGTEE34 pKa = 4.38FQLSAPLAVAICAVFGFAKK53 pKa = 10.65YY54 pKa = 8.25ITAGLLSSMAGLMLGTQSVWNVGIALIFRR83 pKa = 11.84CVVGAVFVLGGNRR96 pKa = 11.84WLTVVLAGPIATITSRR112 pKa = 11.84LVVGGILGNMAIPFMLAAVPGIIYY136 pKa = 8.09TAITVWPLTILLKK149 pKa = 10.68RR150 pKa = 11.84ITEE153 pKa = 4.1RR154 pKa = 11.84GKK156 pKa = 9.85KK157 pKa = 9.77AIVYY161 pKa = 7.51VVQRR165 pKa = 3.91
Molecular weight: 17.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1252127 |
25 |
2803 |
303.5 |
33.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.244 ± 0.05 | 1.155 ± 0.017 |
4.878 ± 0.027 | 6.315 ± 0.038 |
3.888 ± 0.03 | 7.544 ± 0.04 |
1.917 ± 0.019 | 7.037 ± 0.035 |
5.52 ± 0.038 | 10.296 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.636 ± 0.02 | 3.802 ± 0.034 |
3.892 ± 0.023 | 4.113 ± 0.027 |
4.823 ± 0.032 | 5.601 ± 0.036 |
5.5 ± 0.037 | 7.514 ± 0.032 |
0.972 ± 0.014 | 3.352 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
