
Anthoxanthum odoratum amalgavirus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Amalgaviridae; unclassified Amalgaviridae
Average proteome isoelectric point is 6.93
Get precalculated fractions of proteins
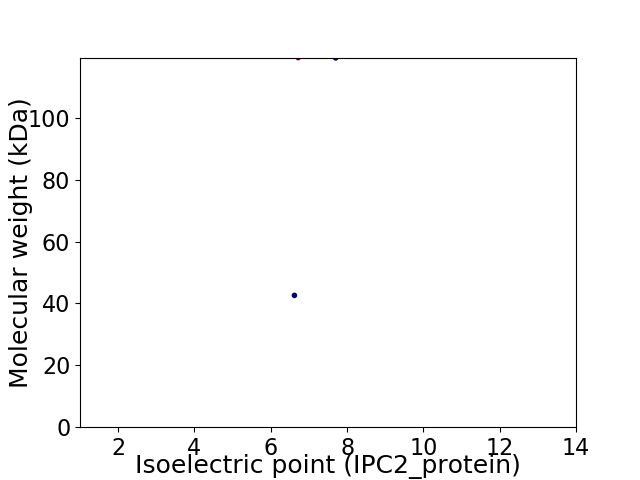
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2V0A0Q4|A0A2V0A0Q4_9VIRU RNA-dependent RNA polymerase OS=Anthoxanthum odoratum amalgavirus 1 OX=2065797 GN=ORF1+ORF2 PE=4 SV=1
MM1 pKa = 7.78AEE3 pKa = 4.05PRR5 pKa = 11.84RR6 pKa = 11.84TFQTAAPTEE15 pKa = 4.09GDD17 pKa = 3.75YY18 pKa = 11.05VANLPADD25 pKa = 3.71EE26 pKa = 5.1DD27 pKa = 3.96AAFVYY32 pKa = 8.93FARR35 pKa = 11.84WVFTTYY41 pKa = 10.76HH42 pKa = 6.61LAAGLLDD49 pKa = 3.97PATYY53 pKa = 10.12RR54 pKa = 11.84PEE56 pKa = 4.71GYY58 pKa = 9.63SDD60 pKa = 3.77KK61 pKa = 11.68DD62 pKa = 3.01MAARR66 pKa = 11.84LRR68 pKa = 11.84FFKK71 pKa = 10.8GKK73 pKa = 9.4EE74 pKa = 3.55ADD76 pKa = 3.78VIDD79 pKa = 4.31TIFAVGVRR87 pKa = 11.84KK88 pKa = 10.29SFFTAKK94 pKa = 10.1EE95 pKa = 3.98SATFEE100 pKa = 3.89QFANFLEE107 pKa = 4.37FLRR110 pKa = 11.84TADD113 pKa = 3.01GAAAVTEE120 pKa = 4.11DD121 pKa = 3.51LRR123 pKa = 11.84RR124 pKa = 11.84SRR126 pKa = 11.84FQAAGKK132 pKa = 10.22GVFNAVEE139 pKa = 4.11ISSVAQLTVQLADD152 pKa = 4.05LEE154 pKa = 4.48LHH156 pKa = 5.46KK157 pKa = 10.52HH158 pKa = 5.18RR159 pKa = 11.84MSDD162 pKa = 3.25ASQKK166 pKa = 10.8RR167 pKa = 11.84IDD169 pKa = 3.98EE170 pKa = 4.52LEE172 pKa = 3.9LLIAKK177 pKa = 9.17EE178 pKa = 4.13KK179 pKa = 10.87EE180 pKa = 4.11SLEE183 pKa = 4.5TKK185 pKa = 10.57LKK187 pKa = 10.42MSDD190 pKa = 3.57EE191 pKa = 4.4EE192 pKa = 4.4FFPASIYY199 pKa = 10.84KK200 pKa = 10.18KK201 pKa = 9.96LNPMKK206 pKa = 10.13LQKK209 pKa = 10.2EE210 pKa = 4.24CWVRR214 pKa = 11.84AKK216 pKa = 10.81ASSTEE221 pKa = 3.93LAAVAVPTNDD231 pKa = 3.4QLADD235 pKa = 3.28ALKK238 pKa = 9.83NFKK241 pKa = 10.83EE242 pKa = 4.31EE243 pKa = 3.8VEE245 pKa = 4.14RR246 pKa = 11.84RR247 pKa = 11.84HH248 pKa = 6.53AVDD251 pKa = 6.31FLAQQNRR258 pKa = 11.84LTALRR263 pKa = 11.84EE264 pKa = 4.01YY265 pKa = 10.68VNKK268 pKa = 10.54KK269 pKa = 9.35ILSFRR274 pKa = 11.84ARR276 pKa = 11.84GKK278 pKa = 10.32YY279 pKa = 9.87RR280 pKa = 11.84IATRR284 pKa = 11.84LGHH287 pKa = 6.24VLAAMDD293 pKa = 4.1GQASGEE299 pKa = 4.17VPPAGAHH306 pKa = 6.42IGDD309 pKa = 4.59GDD311 pKa = 4.05DD312 pKa = 5.33SDD314 pKa = 5.43GEE316 pKa = 4.62DD317 pKa = 4.07YY318 pKa = 11.08IPDD321 pKa = 3.78YY322 pKa = 10.98EE323 pKa = 5.09RR324 pKa = 11.84EE325 pKa = 4.1LSDD328 pKa = 3.48EE329 pKa = 4.24SSGSDD334 pKa = 3.65EE335 pKa = 4.37EE336 pKa = 4.6SGSAGGTPEE345 pKa = 3.9RR346 pKa = 11.84HH347 pKa = 5.36RR348 pKa = 11.84TPTPARR354 pKa = 11.84SRR356 pKa = 11.84RR357 pKa = 11.84PRR359 pKa = 11.84RR360 pKa = 11.84RR361 pKa = 11.84RR362 pKa = 11.84DD363 pKa = 3.25YY364 pKa = 10.44TGLARR369 pKa = 11.84GSAPARR375 pKa = 11.84NTRR378 pKa = 11.84SRR380 pKa = 11.84KK381 pKa = 9.27AA382 pKa = 3.17
MM1 pKa = 7.78AEE3 pKa = 4.05PRR5 pKa = 11.84RR6 pKa = 11.84TFQTAAPTEE15 pKa = 4.09GDD17 pKa = 3.75YY18 pKa = 11.05VANLPADD25 pKa = 3.71EE26 pKa = 5.1DD27 pKa = 3.96AAFVYY32 pKa = 8.93FARR35 pKa = 11.84WVFTTYY41 pKa = 10.76HH42 pKa = 6.61LAAGLLDD49 pKa = 3.97PATYY53 pKa = 10.12RR54 pKa = 11.84PEE56 pKa = 4.71GYY58 pKa = 9.63SDD60 pKa = 3.77KK61 pKa = 11.68DD62 pKa = 3.01MAARR66 pKa = 11.84LRR68 pKa = 11.84FFKK71 pKa = 10.8GKK73 pKa = 9.4EE74 pKa = 3.55ADD76 pKa = 3.78VIDD79 pKa = 4.31TIFAVGVRR87 pKa = 11.84KK88 pKa = 10.29SFFTAKK94 pKa = 10.1EE95 pKa = 3.98SATFEE100 pKa = 3.89QFANFLEE107 pKa = 4.37FLRR110 pKa = 11.84TADD113 pKa = 3.01GAAAVTEE120 pKa = 4.11DD121 pKa = 3.51LRR123 pKa = 11.84RR124 pKa = 11.84SRR126 pKa = 11.84FQAAGKK132 pKa = 10.22GVFNAVEE139 pKa = 4.11ISSVAQLTVQLADD152 pKa = 4.05LEE154 pKa = 4.48LHH156 pKa = 5.46KK157 pKa = 10.52HH158 pKa = 5.18RR159 pKa = 11.84MSDD162 pKa = 3.25ASQKK166 pKa = 10.8RR167 pKa = 11.84IDD169 pKa = 3.98EE170 pKa = 4.52LEE172 pKa = 3.9LLIAKK177 pKa = 9.17EE178 pKa = 4.13KK179 pKa = 10.87EE180 pKa = 4.11SLEE183 pKa = 4.5TKK185 pKa = 10.57LKK187 pKa = 10.42MSDD190 pKa = 3.57EE191 pKa = 4.4EE192 pKa = 4.4FFPASIYY199 pKa = 10.84KK200 pKa = 10.18KK201 pKa = 9.96LNPMKK206 pKa = 10.13LQKK209 pKa = 10.2EE210 pKa = 4.24CWVRR214 pKa = 11.84AKK216 pKa = 10.81ASSTEE221 pKa = 3.93LAAVAVPTNDD231 pKa = 3.4QLADD235 pKa = 3.28ALKK238 pKa = 9.83NFKK241 pKa = 10.83EE242 pKa = 4.31EE243 pKa = 3.8VEE245 pKa = 4.14RR246 pKa = 11.84RR247 pKa = 11.84HH248 pKa = 6.53AVDD251 pKa = 6.31FLAQQNRR258 pKa = 11.84LTALRR263 pKa = 11.84EE264 pKa = 4.01YY265 pKa = 10.68VNKK268 pKa = 10.54KK269 pKa = 9.35ILSFRR274 pKa = 11.84ARR276 pKa = 11.84GKK278 pKa = 10.32YY279 pKa = 9.87RR280 pKa = 11.84IATRR284 pKa = 11.84LGHH287 pKa = 6.24VLAAMDD293 pKa = 4.1GQASGEE299 pKa = 4.17VPPAGAHH306 pKa = 6.42IGDD309 pKa = 4.59GDD311 pKa = 4.05DD312 pKa = 5.33SDD314 pKa = 5.43GEE316 pKa = 4.62DD317 pKa = 4.07YY318 pKa = 11.08IPDD321 pKa = 3.78YY322 pKa = 10.98EE323 pKa = 5.09RR324 pKa = 11.84EE325 pKa = 4.1LSDD328 pKa = 3.48EE329 pKa = 4.24SSGSDD334 pKa = 3.65EE335 pKa = 4.37EE336 pKa = 4.6SGSAGGTPEE345 pKa = 3.9RR346 pKa = 11.84HH347 pKa = 5.36RR348 pKa = 11.84TPTPARR354 pKa = 11.84SRR356 pKa = 11.84RR357 pKa = 11.84PRR359 pKa = 11.84RR360 pKa = 11.84RR361 pKa = 11.84RR362 pKa = 11.84DD363 pKa = 3.25YY364 pKa = 10.44TGLARR369 pKa = 11.84GSAPARR375 pKa = 11.84NTRR378 pKa = 11.84SRR380 pKa = 11.84KK381 pKa = 9.27AA382 pKa = 3.17
Molecular weight: 42.61 kDa
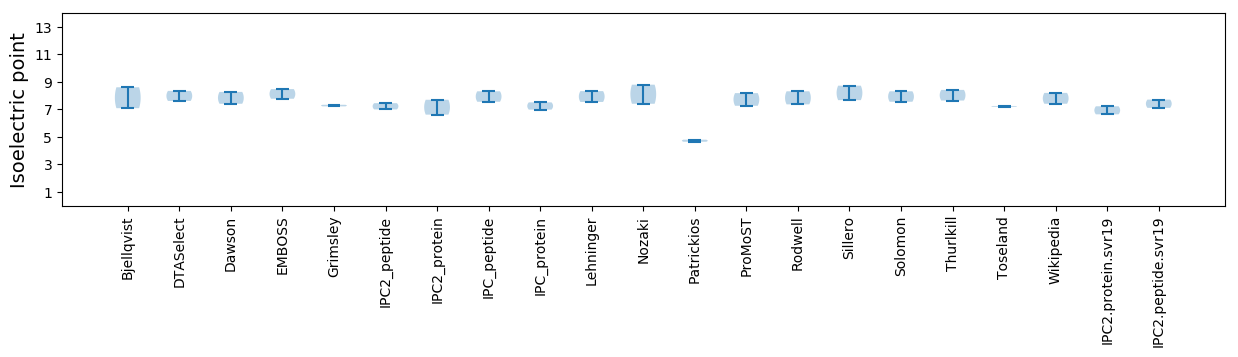
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2V0A0Q4|A0A2V0A0Q4_9VIRU RNA-dependent RNA polymerase OS=Anthoxanthum odoratum amalgavirus 1 OX=2065797 GN=ORF1+ORF2 PE=4 SV=1
MM1 pKa = 7.78AEE3 pKa = 4.05PRR5 pKa = 11.84RR6 pKa = 11.84TFQTAAPTEE15 pKa = 4.09GDD17 pKa = 3.75YY18 pKa = 11.05VANLPADD25 pKa = 3.71EE26 pKa = 5.1DD27 pKa = 3.96AAFVYY32 pKa = 8.93FARR35 pKa = 11.84WVFTTYY41 pKa = 10.76HH42 pKa = 6.61LAAGLLDD49 pKa = 3.97PATYY53 pKa = 10.12RR54 pKa = 11.84PEE56 pKa = 4.71GYY58 pKa = 9.63SDD60 pKa = 3.77KK61 pKa = 11.68DD62 pKa = 3.01MAARR66 pKa = 11.84LRR68 pKa = 11.84FFKK71 pKa = 10.8GKK73 pKa = 9.4EE74 pKa = 3.55ADD76 pKa = 3.78VIDD79 pKa = 4.31TIFAVGVRR87 pKa = 11.84KK88 pKa = 10.29SFFTAKK94 pKa = 10.1EE95 pKa = 3.98SATFEE100 pKa = 3.89QFANFLEE107 pKa = 4.37FLRR110 pKa = 11.84TADD113 pKa = 3.01GAAAVTEE120 pKa = 4.11DD121 pKa = 3.51LRR123 pKa = 11.84RR124 pKa = 11.84SRR126 pKa = 11.84FQAAGKK132 pKa = 10.22GVFNAVEE139 pKa = 4.11ISSVAQLTVQLADD152 pKa = 4.05LEE154 pKa = 4.48LHH156 pKa = 5.46KK157 pKa = 10.52HH158 pKa = 5.18RR159 pKa = 11.84MSDD162 pKa = 3.25ASQKK166 pKa = 10.8RR167 pKa = 11.84IDD169 pKa = 3.98EE170 pKa = 4.52LEE172 pKa = 3.9LLIAKK177 pKa = 9.17EE178 pKa = 4.13KK179 pKa = 10.87EE180 pKa = 4.11SLEE183 pKa = 4.5TKK185 pKa = 10.57LKK187 pKa = 10.42MSDD190 pKa = 3.57EE191 pKa = 4.4EE192 pKa = 4.4FFPASIYY199 pKa = 10.84KK200 pKa = 10.18KK201 pKa = 9.96LNPMKK206 pKa = 10.13LQKK209 pKa = 10.2EE210 pKa = 4.24CWVRR214 pKa = 11.84AKK216 pKa = 10.81ASSTEE221 pKa = 3.93LAAVAVPTNDD231 pKa = 3.4QLADD235 pKa = 3.28ALKK238 pKa = 9.83NFKK241 pKa = 10.83EE242 pKa = 4.31EE243 pKa = 3.8VEE245 pKa = 4.14RR246 pKa = 11.84RR247 pKa = 11.84HH248 pKa = 6.53AVDD251 pKa = 6.31FLAQQNRR258 pKa = 11.84LTALRR263 pKa = 11.84EE264 pKa = 4.01YY265 pKa = 10.68VNKK268 pKa = 10.52KK269 pKa = 9.13ILSFVLVEE277 pKa = 4.29NTGSRR282 pKa = 11.84PALDD286 pKa = 3.98TSWLPWTAKK295 pKa = 10.54LLEE298 pKa = 4.85KK299 pKa = 10.89YY300 pKa = 9.01PLPEE304 pKa = 5.16RR305 pKa = 11.84ISATAMIPMGRR316 pKa = 11.84TTSPTTSVSCRR327 pKa = 11.84TNLLEE332 pKa = 4.54VMRR335 pKa = 11.84SPALLAVRR343 pKa = 11.84QRR345 pKa = 11.84GTGPPPRR352 pKa = 11.84LGAGVPAAVGTTPVLQGGARR372 pKa = 11.84LHH374 pKa = 6.23VIRR377 pKa = 11.84APVRR381 pKa = 11.84PDD383 pKa = 2.86RR384 pKa = 11.84RR385 pKa = 11.84GIPYY389 pKa = 10.3ARR391 pKa = 11.84SKK393 pKa = 10.32WEE395 pKa = 3.79AGVRR399 pKa = 11.84KK400 pKa = 9.86IIGGGEE406 pKa = 3.87LSDD409 pKa = 3.45WQAASSKK416 pKa = 9.72VRR418 pKa = 11.84CGGNASDD425 pKa = 5.64ALLLLADD432 pKa = 4.48ASDD435 pKa = 3.86RR436 pKa = 11.84LPGKK440 pKa = 6.8TLRR443 pKa = 11.84GLFTLRR449 pKa = 11.84GARR452 pKa = 11.84DD453 pKa = 3.66ALRR456 pKa = 11.84LPSGLSVPDD465 pKa = 4.25GRR467 pKa = 11.84ACCNIKK473 pKa = 10.27HH474 pKa = 6.09FNVEE478 pKa = 3.64ATAGPFLRR486 pKa = 11.84AFGVKK491 pKa = 9.8KK492 pKa = 10.6KK493 pKa = 10.76EE494 pKa = 4.09SLEE497 pKa = 3.83QLLGDD502 pKa = 4.22FVWEE506 pKa = 4.52CFDD509 pKa = 6.04DD510 pKa = 4.87FATNDD515 pKa = 2.99ADD517 pKa = 3.83IRR519 pKa = 11.84RR520 pKa = 11.84LPFFGMRR527 pKa = 11.84IGFRR531 pKa = 11.84TKK533 pKa = 10.36LLKK536 pKa = 10.35KK537 pKa = 10.41SEE539 pKa = 4.32MLTKK543 pKa = 10.15IRR545 pKa = 11.84DD546 pKa = 3.9FKK548 pKa = 11.17PLGRR552 pKa = 11.84CVMMLDD558 pKa = 5.12AIEE561 pKa = 4.41QFCSSPLYY569 pKa = 10.56NVLSKK574 pKa = 10.97LSADD578 pKa = 3.55ALKK581 pKa = 11.07DD582 pKa = 3.45PMSGFRR588 pKa = 11.84NTAVRR593 pKa = 11.84ASSDD597 pKa = 2.64WSYY600 pKa = 9.73MWEE603 pKa = 4.09EE604 pKa = 3.54IKK606 pKa = 10.33EE607 pKa = 4.24AKK609 pKa = 10.17VCMEE613 pKa = 4.68LDD615 pKa = 2.81WSKK618 pKa = 10.93FDD620 pKa = 4.73RR621 pKa = 11.84EE622 pKa = 4.11RR623 pKa = 11.84PRR625 pKa = 11.84CDD627 pKa = 3.33LEE629 pKa = 4.55FMVDD633 pKa = 3.59VVISCFAPKK642 pKa = 10.29NEE644 pKa = 3.88RR645 pKa = 11.84EE646 pKa = 3.86QRR648 pKa = 11.84LLRR651 pKa = 11.84GYY653 pKa = 6.91EE654 pKa = 3.63VCMRR658 pKa = 11.84RR659 pKa = 11.84AIVEE663 pKa = 4.18RR664 pKa = 11.84VALLDD669 pKa = 4.29DD670 pKa = 4.4GALFEE675 pKa = 5.2IDD677 pKa = 3.61GMVPSGSLWTGWLDD691 pKa = 3.31TALNILYY698 pKa = 10.4LSAALSEE705 pKa = 4.86AGFSSYY711 pKa = 7.45MARR714 pKa = 11.84PKK716 pKa = 10.74CAGDD720 pKa = 3.9DD721 pKa = 3.7NLTLFMEE728 pKa = 4.81DD729 pKa = 3.42VPDD732 pKa = 3.63GRR734 pKa = 11.84LIQVRR739 pKa = 11.84GLLNDD744 pKa = 3.6WFLAGIKK751 pKa = 10.57DD752 pKa = 3.58EE753 pKa = 5.12DD754 pKa = 3.99FAITRR759 pKa = 11.84APFHH763 pKa = 6.8VEE765 pKa = 4.08TYY767 pKa = 10.35QAVFPPGTDD776 pKa = 3.29LTKK779 pKa = 10.06GTSKK783 pKa = 10.63IIDD786 pKa = 3.18QCEE789 pKa = 3.68WVRR792 pKa = 11.84FEE794 pKa = 4.82GQLQINQEE802 pKa = 4.01QGLSHH807 pKa = 6.82RR808 pKa = 11.84WQYY811 pKa = 10.66RR812 pKa = 11.84FKK814 pKa = 10.9GKK816 pKa = 9.57PKK818 pKa = 10.28FLSCYY823 pKa = 8.28WLADD827 pKa = 3.34GRR829 pKa = 11.84PIRR832 pKa = 11.84PASDD836 pKa = 3.13NVEE839 pKa = 3.83KK840 pKa = 10.78LLYY843 pKa = 10.0PEE845 pKa = 5.91GIHH848 pKa = 7.08KK849 pKa = 10.59SVEE852 pKa = 4.29DD853 pKa = 4.09YY854 pKa = 11.09ISACLAMVVDD864 pKa = 4.82NPFNQHH870 pKa = 5.29NVNHH874 pKa = 6.16MKK876 pKa = 10.09HH877 pKa = 6.55RR878 pKa = 11.84YY879 pKa = 9.24LIAQQIKK886 pKa = 9.37RR887 pKa = 11.84ILVTGIPDD895 pKa = 3.77RR896 pKa = 11.84LVLALARR903 pKa = 11.84IRR905 pKa = 11.84PDD907 pKa = 3.29GEE909 pKa = 3.6EE910 pKa = 4.66DD911 pKa = 2.97IPYY914 pKa = 10.64PMIAPWRR921 pKa = 11.84RR922 pKa = 11.84FKK924 pKa = 10.93EE925 pKa = 4.18YY926 pKa = 10.19IDD928 pKa = 4.17LDD930 pKa = 3.74SYY932 pKa = 11.77EE933 pKa = 5.42PVQQWIKK940 pKa = 10.9EE941 pKa = 3.92FDD943 pKa = 3.78DD944 pKa = 4.77FVAGISGLYY953 pKa = 9.33VRR955 pKa = 11.84STTGGIDD962 pKa = 2.88AYY964 pKa = 10.26KK965 pKa = 10.72FMDD968 pKa = 5.57FIRR971 pKa = 11.84GDD973 pKa = 3.5AVIGEE978 pKa = 4.5GQWGNEE984 pKa = 3.45MDD986 pKa = 2.9RR987 pKa = 11.84WIRR990 pKa = 11.84FITEE994 pKa = 4.03HH995 pKa = 5.98PVSRR999 pKa = 11.84SLRR1002 pKa = 11.84KK1003 pKa = 9.53ARR1005 pKa = 11.84RR1006 pKa = 11.84HH1007 pKa = 5.56NPASVPSAQVNQYY1020 pKa = 7.03YY1021 pKa = 9.79QRR1023 pKa = 11.84AAEE1026 pKa = 4.06GLNVYY1031 pKa = 9.69RR1032 pKa = 11.84QQLVNHH1038 pKa = 6.83RR1039 pKa = 11.84LDD1041 pKa = 3.01SSRR1044 pKa = 11.84EE1045 pKa = 3.72YY1046 pKa = 11.1GLWVSDD1052 pKa = 3.43LLRR1055 pKa = 11.84RR1056 pKa = 3.93
MM1 pKa = 7.78AEE3 pKa = 4.05PRR5 pKa = 11.84RR6 pKa = 11.84TFQTAAPTEE15 pKa = 4.09GDD17 pKa = 3.75YY18 pKa = 11.05VANLPADD25 pKa = 3.71EE26 pKa = 5.1DD27 pKa = 3.96AAFVYY32 pKa = 8.93FARR35 pKa = 11.84WVFTTYY41 pKa = 10.76HH42 pKa = 6.61LAAGLLDD49 pKa = 3.97PATYY53 pKa = 10.12RR54 pKa = 11.84PEE56 pKa = 4.71GYY58 pKa = 9.63SDD60 pKa = 3.77KK61 pKa = 11.68DD62 pKa = 3.01MAARR66 pKa = 11.84LRR68 pKa = 11.84FFKK71 pKa = 10.8GKK73 pKa = 9.4EE74 pKa = 3.55ADD76 pKa = 3.78VIDD79 pKa = 4.31TIFAVGVRR87 pKa = 11.84KK88 pKa = 10.29SFFTAKK94 pKa = 10.1EE95 pKa = 3.98SATFEE100 pKa = 3.89QFANFLEE107 pKa = 4.37FLRR110 pKa = 11.84TADD113 pKa = 3.01GAAAVTEE120 pKa = 4.11DD121 pKa = 3.51LRR123 pKa = 11.84RR124 pKa = 11.84SRR126 pKa = 11.84FQAAGKK132 pKa = 10.22GVFNAVEE139 pKa = 4.11ISSVAQLTVQLADD152 pKa = 4.05LEE154 pKa = 4.48LHH156 pKa = 5.46KK157 pKa = 10.52HH158 pKa = 5.18RR159 pKa = 11.84MSDD162 pKa = 3.25ASQKK166 pKa = 10.8RR167 pKa = 11.84IDD169 pKa = 3.98EE170 pKa = 4.52LEE172 pKa = 3.9LLIAKK177 pKa = 9.17EE178 pKa = 4.13KK179 pKa = 10.87EE180 pKa = 4.11SLEE183 pKa = 4.5TKK185 pKa = 10.57LKK187 pKa = 10.42MSDD190 pKa = 3.57EE191 pKa = 4.4EE192 pKa = 4.4FFPASIYY199 pKa = 10.84KK200 pKa = 10.18KK201 pKa = 9.96LNPMKK206 pKa = 10.13LQKK209 pKa = 10.2EE210 pKa = 4.24CWVRR214 pKa = 11.84AKK216 pKa = 10.81ASSTEE221 pKa = 3.93LAAVAVPTNDD231 pKa = 3.4QLADD235 pKa = 3.28ALKK238 pKa = 9.83NFKK241 pKa = 10.83EE242 pKa = 4.31EE243 pKa = 3.8VEE245 pKa = 4.14RR246 pKa = 11.84RR247 pKa = 11.84HH248 pKa = 6.53AVDD251 pKa = 6.31FLAQQNRR258 pKa = 11.84LTALRR263 pKa = 11.84EE264 pKa = 4.01YY265 pKa = 10.68VNKK268 pKa = 10.52KK269 pKa = 9.13ILSFVLVEE277 pKa = 4.29NTGSRR282 pKa = 11.84PALDD286 pKa = 3.98TSWLPWTAKK295 pKa = 10.54LLEE298 pKa = 4.85KK299 pKa = 10.89YY300 pKa = 9.01PLPEE304 pKa = 5.16RR305 pKa = 11.84ISATAMIPMGRR316 pKa = 11.84TTSPTTSVSCRR327 pKa = 11.84TNLLEE332 pKa = 4.54VMRR335 pKa = 11.84SPALLAVRR343 pKa = 11.84QRR345 pKa = 11.84GTGPPPRR352 pKa = 11.84LGAGVPAAVGTTPVLQGGARR372 pKa = 11.84LHH374 pKa = 6.23VIRR377 pKa = 11.84APVRR381 pKa = 11.84PDD383 pKa = 2.86RR384 pKa = 11.84RR385 pKa = 11.84GIPYY389 pKa = 10.3ARR391 pKa = 11.84SKK393 pKa = 10.32WEE395 pKa = 3.79AGVRR399 pKa = 11.84KK400 pKa = 9.86IIGGGEE406 pKa = 3.87LSDD409 pKa = 3.45WQAASSKK416 pKa = 9.72VRR418 pKa = 11.84CGGNASDD425 pKa = 5.64ALLLLADD432 pKa = 4.48ASDD435 pKa = 3.86RR436 pKa = 11.84LPGKK440 pKa = 6.8TLRR443 pKa = 11.84GLFTLRR449 pKa = 11.84GARR452 pKa = 11.84DD453 pKa = 3.66ALRR456 pKa = 11.84LPSGLSVPDD465 pKa = 4.25GRR467 pKa = 11.84ACCNIKK473 pKa = 10.27HH474 pKa = 6.09FNVEE478 pKa = 3.64ATAGPFLRR486 pKa = 11.84AFGVKK491 pKa = 9.8KK492 pKa = 10.6KK493 pKa = 10.76EE494 pKa = 4.09SLEE497 pKa = 3.83QLLGDD502 pKa = 4.22FVWEE506 pKa = 4.52CFDD509 pKa = 6.04DD510 pKa = 4.87FATNDD515 pKa = 2.99ADD517 pKa = 3.83IRR519 pKa = 11.84RR520 pKa = 11.84LPFFGMRR527 pKa = 11.84IGFRR531 pKa = 11.84TKK533 pKa = 10.36LLKK536 pKa = 10.35KK537 pKa = 10.41SEE539 pKa = 4.32MLTKK543 pKa = 10.15IRR545 pKa = 11.84DD546 pKa = 3.9FKK548 pKa = 11.17PLGRR552 pKa = 11.84CVMMLDD558 pKa = 5.12AIEE561 pKa = 4.41QFCSSPLYY569 pKa = 10.56NVLSKK574 pKa = 10.97LSADD578 pKa = 3.55ALKK581 pKa = 11.07DD582 pKa = 3.45PMSGFRR588 pKa = 11.84NTAVRR593 pKa = 11.84ASSDD597 pKa = 2.64WSYY600 pKa = 9.73MWEE603 pKa = 4.09EE604 pKa = 3.54IKK606 pKa = 10.33EE607 pKa = 4.24AKK609 pKa = 10.17VCMEE613 pKa = 4.68LDD615 pKa = 2.81WSKK618 pKa = 10.93FDD620 pKa = 4.73RR621 pKa = 11.84EE622 pKa = 4.11RR623 pKa = 11.84PRR625 pKa = 11.84CDD627 pKa = 3.33LEE629 pKa = 4.55FMVDD633 pKa = 3.59VVISCFAPKK642 pKa = 10.29NEE644 pKa = 3.88RR645 pKa = 11.84EE646 pKa = 3.86QRR648 pKa = 11.84LLRR651 pKa = 11.84GYY653 pKa = 6.91EE654 pKa = 3.63VCMRR658 pKa = 11.84RR659 pKa = 11.84AIVEE663 pKa = 4.18RR664 pKa = 11.84VALLDD669 pKa = 4.29DD670 pKa = 4.4GALFEE675 pKa = 5.2IDD677 pKa = 3.61GMVPSGSLWTGWLDD691 pKa = 3.31TALNILYY698 pKa = 10.4LSAALSEE705 pKa = 4.86AGFSSYY711 pKa = 7.45MARR714 pKa = 11.84PKK716 pKa = 10.74CAGDD720 pKa = 3.9DD721 pKa = 3.7NLTLFMEE728 pKa = 4.81DD729 pKa = 3.42VPDD732 pKa = 3.63GRR734 pKa = 11.84LIQVRR739 pKa = 11.84GLLNDD744 pKa = 3.6WFLAGIKK751 pKa = 10.57DD752 pKa = 3.58EE753 pKa = 5.12DD754 pKa = 3.99FAITRR759 pKa = 11.84APFHH763 pKa = 6.8VEE765 pKa = 4.08TYY767 pKa = 10.35QAVFPPGTDD776 pKa = 3.29LTKK779 pKa = 10.06GTSKK783 pKa = 10.63IIDD786 pKa = 3.18QCEE789 pKa = 3.68WVRR792 pKa = 11.84FEE794 pKa = 4.82GQLQINQEE802 pKa = 4.01QGLSHH807 pKa = 6.82RR808 pKa = 11.84WQYY811 pKa = 10.66RR812 pKa = 11.84FKK814 pKa = 10.9GKK816 pKa = 9.57PKK818 pKa = 10.28FLSCYY823 pKa = 8.28WLADD827 pKa = 3.34GRR829 pKa = 11.84PIRR832 pKa = 11.84PASDD836 pKa = 3.13NVEE839 pKa = 3.83KK840 pKa = 10.78LLYY843 pKa = 10.0PEE845 pKa = 5.91GIHH848 pKa = 7.08KK849 pKa = 10.59SVEE852 pKa = 4.29DD853 pKa = 4.09YY854 pKa = 11.09ISACLAMVVDD864 pKa = 4.82NPFNQHH870 pKa = 5.29NVNHH874 pKa = 6.16MKK876 pKa = 10.09HH877 pKa = 6.55RR878 pKa = 11.84YY879 pKa = 9.24LIAQQIKK886 pKa = 9.37RR887 pKa = 11.84ILVTGIPDD895 pKa = 3.77RR896 pKa = 11.84LVLALARR903 pKa = 11.84IRR905 pKa = 11.84PDD907 pKa = 3.29GEE909 pKa = 3.6EE910 pKa = 4.66DD911 pKa = 2.97IPYY914 pKa = 10.64PMIAPWRR921 pKa = 11.84RR922 pKa = 11.84FKK924 pKa = 10.93EE925 pKa = 4.18YY926 pKa = 10.19IDD928 pKa = 4.17LDD930 pKa = 3.74SYY932 pKa = 11.77EE933 pKa = 5.42PVQQWIKK940 pKa = 10.9EE941 pKa = 3.92FDD943 pKa = 3.78DD944 pKa = 4.77FVAGISGLYY953 pKa = 9.33VRR955 pKa = 11.84STTGGIDD962 pKa = 2.88AYY964 pKa = 10.26KK965 pKa = 10.72FMDD968 pKa = 5.57FIRR971 pKa = 11.84GDD973 pKa = 3.5AVIGEE978 pKa = 4.5GQWGNEE984 pKa = 3.45MDD986 pKa = 2.9RR987 pKa = 11.84WIRR990 pKa = 11.84FITEE994 pKa = 4.03HH995 pKa = 5.98PVSRR999 pKa = 11.84SLRR1002 pKa = 11.84KK1003 pKa = 9.53ARR1005 pKa = 11.84RR1006 pKa = 11.84HH1007 pKa = 5.56NPASVPSAQVNQYY1020 pKa = 7.03YY1021 pKa = 9.79QRR1023 pKa = 11.84AAEE1026 pKa = 4.06GLNVYY1031 pKa = 9.69RR1032 pKa = 11.84QQLVNHH1038 pKa = 6.83RR1039 pKa = 11.84LDD1041 pKa = 3.01SSRR1044 pKa = 11.84EE1045 pKa = 3.72YY1046 pKa = 11.1GLWVSDD1052 pKa = 3.43LLRR1055 pKa = 11.84RR1056 pKa = 3.93
Molecular weight: 119.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1438 |
382 |
1056 |
719.0 |
81.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.848 ± 1.491 | 1.182 ± 0.454 |
6.676 ± 0.193 | 6.885 ± 0.735 |
5.076 ± 0.078 | 6.05 ± 0.143 |
1.53 ± 0.149 | 3.964 ± 0.663 |
5.563 ± 0.226 | 9.458 ± 0.791 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.156 ± 0.288 | 2.782 ± 0.21 |
4.798 ± 0.301 | 3.199 ± 0.157 |
8.623 ± 0.395 | 6.12 ± 0.209 |
4.798 ± 0.473 | 5.911 ± 0.462 |
1.599 ± 0.53 | 2.782 ± 0.048 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
