
Fulvivirga sp. M361
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Fulvivirgaceae; Fulvivirga; unclassified Fulvivirga
Average proteome isoelectric point is 6.28
Get precalculated fractions of proteins
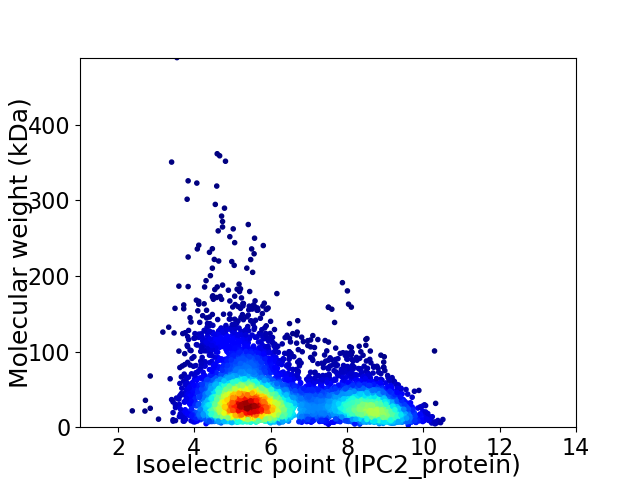
Virtual 2D-PAGE plot for 6272 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A553FFQ2|A0A553FFQ2_9BACT NADH-quinone oxidoreductase subunit H OS=Fulvivirga sp. M361 OX=2594266 GN=nuoH PE=3 SV=1
MM1 pKa = 7.32SHH3 pKa = 5.92TEE5 pKa = 3.78LRR7 pKa = 11.84RR8 pKa = 11.84VCLLLIFVVGSFSNLLAQSYY28 pKa = 10.94SGINWYY34 pKa = 10.1FGNNDD39 pKa = 2.5KK40 pKa = 10.91SIRR43 pKa = 11.84FTRR46 pKa = 11.84PTLAPDD52 pKa = 3.64TLSLPNILGAGGGAVATDD70 pKa = 5.3AITGEE75 pKa = 3.47IWFYY79 pKa = 10.36TDD81 pKa = 4.77GINVYY86 pKa = 10.76DD87 pKa = 3.68RR88 pKa = 11.84TDD90 pKa = 3.53FQLATTVDD98 pKa = 3.62GSTTRR103 pKa = 11.84NQSVAICEE111 pKa = 4.11NPANSDD117 pKa = 3.4QYY119 pKa = 12.0FLFTINNAGAIVQSIFDD136 pKa = 3.73KK137 pKa = 10.4TQFRR141 pKa = 11.84PGGAFPDD148 pKa = 3.92SPEE151 pKa = 4.32GNFTSLNNAVAPVTGPLSEE170 pKa = 4.13GMIILSNDD178 pKa = 3.09ALDD181 pKa = 4.38GFWLITQTQGGDD193 pKa = 3.26INVTEE198 pKa = 4.29IDD200 pKa = 3.47GTGTFNTTTFATTLTQIDD218 pKa = 4.21NFSFHH223 pKa = 7.07PSSNQLAAASAQAGEE238 pKa = 4.42GVEE241 pKa = 5.68LFDD244 pKa = 5.78FDD246 pKa = 4.62PATGAVTASATSLAAITFNGVYY268 pKa = 9.0DD269 pKa = 4.19TEE271 pKa = 4.62WNNSGQYY278 pKa = 10.57LYY280 pKa = 11.28ASGNFGQPEE289 pKa = 4.05DD290 pKa = 3.68SLIRR294 pKa = 11.84IDD296 pKa = 4.99LNAVPPTVEE305 pKa = 4.21NIQTFGVTNSFGIQIAPDD323 pKa = 3.25STIYY327 pKa = 10.79HH328 pKa = 6.87LFEE331 pKa = 6.02DD332 pKa = 3.95PAGAFRR338 pKa = 11.84VGRR341 pKa = 11.84INDD344 pKa = 3.96PDD346 pKa = 3.84TSDD349 pKa = 3.43VAQVLYY355 pKa = 10.5DD356 pKa = 3.77PRR358 pKa = 11.84PFDD361 pKa = 3.76VPDD364 pKa = 4.67FQAEE368 pKa = 4.14QFPSFLPLIDD378 pKa = 4.78PDD380 pKa = 3.79FTLDD384 pKa = 3.53FSFFGDD390 pKa = 3.97CQNEE394 pKa = 3.98PVLFFPEE401 pKa = 4.07ISINPDD407 pKa = 2.74SVRR410 pKa = 11.84WDD412 pKa = 3.5FAGEE416 pKa = 4.11GNSASLAPSFTFSDD430 pKa = 3.67AAGSPFTVTLVAFINGQPRR449 pKa = 11.84AASQQVTINPFDD461 pKa = 3.48ISITLVSDD469 pKa = 3.32TTFCRR474 pKa = 11.84EE475 pKa = 4.13DD476 pKa = 3.65FPPPFGTNGTASVEE490 pKa = 4.11AEE492 pKa = 3.69ISGTPTSVVWSNGQTGNILVPDD514 pKa = 3.75STGFYY519 pKa = 10.46YY520 pKa = 10.83VVATDD525 pKa = 3.6ATGCSTYY532 pKa = 11.16AGVQVNTYY540 pKa = 10.47GEE542 pKa = 3.78QDD544 pKa = 3.1QRR546 pKa = 11.84GNIWYY551 pKa = 9.16FGNNAGIDD559 pKa = 4.07FNPPPPTEE567 pKa = 4.98AIPFGDD573 pKa = 3.18ASIYY577 pKa = 10.94NGGNQLRR584 pKa = 11.84MPEE587 pKa = 4.01GCAIYY592 pKa = 10.06CDD594 pKa = 4.78RR595 pKa = 11.84NGQPLFYY602 pKa = 10.46SDD604 pKa = 3.74GVDD607 pKa = 3.55VFDD610 pKa = 6.22RR611 pKa = 11.84EE612 pKa = 4.62ANLLTPANKK621 pKa = 9.59IGGTAGISQGSSQSVLVLPFPGDD644 pKa = 3.23EE645 pKa = 4.01TLFYY649 pKa = 11.08LFTTNEE655 pKa = 4.0VYY657 pKa = 10.74NPSGSYY663 pKa = 8.19EE664 pKa = 3.75LRR666 pKa = 11.84YY667 pKa = 10.3SVFDD671 pKa = 3.94LKK673 pKa = 11.31LNNGLGGMVLDD684 pKa = 4.14QNGEE688 pKa = 3.68ISTILCSDD696 pKa = 3.21VTEE699 pKa = 4.04RR700 pKa = 11.84LAGNEE705 pKa = 3.39NWVIVHH711 pKa = 6.39EE712 pKa = 4.5LGNNNFRR719 pKa = 11.84AYY721 pKa = 9.39PVLGDD726 pKa = 4.21GIGTPVISNIGAIHH740 pKa = 6.04NSEE743 pKa = 4.23LEE745 pKa = 3.87SRR747 pKa = 11.84GYY749 pKa = 9.65MKK751 pKa = 11.07LNGNSNQLAVAYY763 pKa = 10.59SEE765 pKa = 4.27STNNNFVEE773 pKa = 5.98LFDD776 pKa = 4.64FDD778 pKa = 3.97NATGEE783 pKa = 4.24VSNYY787 pKa = 10.01LQLDD791 pKa = 3.87FNNPVSSAGPVSGQVYY807 pKa = 10.37GIEE810 pKa = 5.12FGNNDD815 pKa = 3.31NNLFASLIGTPSEE828 pKa = 3.72IFLWHH833 pKa = 6.72VDD835 pKa = 3.13TTTIAGNITDD845 pKa = 3.66PTYY848 pKa = 10.69IRR850 pKa = 11.84DD851 pKa = 3.66SVEE854 pKa = 4.12VVDD857 pKa = 6.22LSSLPATDD865 pKa = 5.2SIGALQLGPDD875 pKa = 3.56GQMYY879 pKa = 10.2GAIQNQQFTVTIGNTGTRR897 pKa = 11.84FGQPVPVANTFGLAAGTSSQLGLPNFIQFSGSSSQNASLAVEE939 pKa = 4.82SGCIDD944 pKa = 3.31QPLTFTVNNPLNPNLEE960 pKa = 4.15IYY962 pKa = 9.97FVSIFPEE969 pKa = 4.4GDD971 pKa = 3.07EE972 pKa = 4.68SNVLVGPIQLTQDD985 pKa = 3.35NPDD988 pKa = 3.45FTFSLPDD995 pKa = 3.75PGNYY999 pKa = 8.74VARR1002 pKa = 11.84FFIDD1006 pKa = 3.34QQCIPQQLITDD1017 pKa = 4.14SDD1019 pKa = 3.71QLFAINPLPTVSNIAVTDD1037 pKa = 3.69PTGCGLPDD1045 pKa = 3.83GSAIVTFSTTGNITYY1060 pKa = 10.07SVSGPVSYY1068 pKa = 10.57PDD1070 pKa = 3.18QTVTTTGSTNVTIPALSAGFYY1091 pKa = 9.42TLNAAFDD1098 pKa = 4.12TGCSEE1103 pKa = 4.21TFTFILNDD1111 pKa = 3.66PVPYY1115 pKa = 9.86TLAAVQSQGSDD1126 pKa = 3.21CDD1128 pKa = 3.89DD1129 pKa = 3.44QNGEE1133 pKa = 4.03ITVTLSPLAQVPAAYY1148 pKa = 9.96DD1149 pKa = 3.24WVLNEE1154 pKa = 4.02QSTGTTVRR1162 pKa = 11.84TGNQADD1168 pKa = 4.51DD1169 pKa = 4.39PDD1171 pKa = 4.71SDD1173 pKa = 4.12GVFVITGVSTGNYY1186 pKa = 9.49FLEE1189 pKa = 4.53LTDD1192 pKa = 3.63NAGCISTVNVSITPPPSITLTLPDD1216 pKa = 4.44QFFQACDD1223 pKa = 3.1VDD1225 pKa = 4.96EE1226 pKa = 4.31IVIPITTNSNSLVEE1240 pKa = 4.77LLSQTGDD1247 pKa = 3.19LLTNFRR1253 pKa = 11.84VIGQNDD1259 pKa = 4.15SIAITNPGISGEE1271 pKa = 4.09PIIFGIRR1278 pKa = 11.84ALGEE1282 pKa = 4.21DD1283 pKa = 5.1PISCDD1288 pKa = 3.09TTVVIQVIFDD1298 pKa = 3.62NSSPNPFDD1306 pKa = 4.31SRR1308 pKa = 11.84YY1309 pKa = 7.39TLCPFEE1315 pKa = 4.76SFEE1318 pKa = 4.29SGRR1321 pKa = 11.84QVVMLEE1327 pKa = 4.08NPPSGFQSARR1337 pKa = 11.84WFDD1340 pKa = 3.87RR1341 pKa = 11.84NSVEE1345 pKa = 4.29IEE1347 pKa = 4.09RR1348 pKa = 11.84INASGASNVTGYY1360 pKa = 10.62RR1361 pKa = 11.84FSQAGDD1367 pKa = 3.68SIIIEE1372 pKa = 3.99IAQPISVEE1380 pKa = 3.82LTNAFGCPTVASIDD1394 pKa = 3.39IVQDD1398 pKa = 3.19CKK1400 pKa = 11.31GRR1402 pKa = 11.84INAPSAFYY1410 pKa = 10.22PNSLIPRR1417 pKa = 11.84NRR1419 pKa = 11.84QFVVFPFLILPDD1431 pKa = 4.31DD1432 pKa = 3.73FQIYY1436 pKa = 9.96IFNRR1440 pKa = 11.84WGEE1443 pKa = 4.14MVFQSDD1449 pKa = 3.26NLQFMRR1455 pKa = 11.84EE1456 pKa = 4.01QGWNGGYY1463 pKa = 10.7DD1464 pKa = 3.84NDD1466 pKa = 4.66PGRR1469 pKa = 11.84PVPGGGYY1476 pKa = 10.37AYY1478 pKa = 10.23RR1479 pKa = 11.84INYY1482 pKa = 8.73KK1483 pKa = 9.78SQFEE1487 pKa = 3.92PDD1489 pKa = 3.16EE1490 pKa = 4.71LKK1492 pKa = 10.47EE1493 pKa = 3.95QRR1495 pKa = 11.84GGVTLIRR1502 pKa = 4.51
MM1 pKa = 7.32SHH3 pKa = 5.92TEE5 pKa = 3.78LRR7 pKa = 11.84RR8 pKa = 11.84VCLLLIFVVGSFSNLLAQSYY28 pKa = 10.94SGINWYY34 pKa = 10.1FGNNDD39 pKa = 2.5KK40 pKa = 10.91SIRR43 pKa = 11.84FTRR46 pKa = 11.84PTLAPDD52 pKa = 3.64TLSLPNILGAGGGAVATDD70 pKa = 5.3AITGEE75 pKa = 3.47IWFYY79 pKa = 10.36TDD81 pKa = 4.77GINVYY86 pKa = 10.76DD87 pKa = 3.68RR88 pKa = 11.84TDD90 pKa = 3.53FQLATTVDD98 pKa = 3.62GSTTRR103 pKa = 11.84NQSVAICEE111 pKa = 4.11NPANSDD117 pKa = 3.4QYY119 pKa = 12.0FLFTINNAGAIVQSIFDD136 pKa = 3.73KK137 pKa = 10.4TQFRR141 pKa = 11.84PGGAFPDD148 pKa = 3.92SPEE151 pKa = 4.32GNFTSLNNAVAPVTGPLSEE170 pKa = 4.13GMIILSNDD178 pKa = 3.09ALDD181 pKa = 4.38GFWLITQTQGGDD193 pKa = 3.26INVTEE198 pKa = 4.29IDD200 pKa = 3.47GTGTFNTTTFATTLTQIDD218 pKa = 4.21NFSFHH223 pKa = 7.07PSSNQLAAASAQAGEE238 pKa = 4.42GVEE241 pKa = 5.68LFDD244 pKa = 5.78FDD246 pKa = 4.62PATGAVTASATSLAAITFNGVYY268 pKa = 9.0DD269 pKa = 4.19TEE271 pKa = 4.62WNNSGQYY278 pKa = 10.57LYY280 pKa = 11.28ASGNFGQPEE289 pKa = 4.05DD290 pKa = 3.68SLIRR294 pKa = 11.84IDD296 pKa = 4.99LNAVPPTVEE305 pKa = 4.21NIQTFGVTNSFGIQIAPDD323 pKa = 3.25STIYY327 pKa = 10.79HH328 pKa = 6.87LFEE331 pKa = 6.02DD332 pKa = 3.95PAGAFRR338 pKa = 11.84VGRR341 pKa = 11.84INDD344 pKa = 3.96PDD346 pKa = 3.84TSDD349 pKa = 3.43VAQVLYY355 pKa = 10.5DD356 pKa = 3.77PRR358 pKa = 11.84PFDD361 pKa = 3.76VPDD364 pKa = 4.67FQAEE368 pKa = 4.14QFPSFLPLIDD378 pKa = 4.78PDD380 pKa = 3.79FTLDD384 pKa = 3.53FSFFGDD390 pKa = 3.97CQNEE394 pKa = 3.98PVLFFPEE401 pKa = 4.07ISINPDD407 pKa = 2.74SVRR410 pKa = 11.84WDD412 pKa = 3.5FAGEE416 pKa = 4.11GNSASLAPSFTFSDD430 pKa = 3.67AAGSPFTVTLVAFINGQPRR449 pKa = 11.84AASQQVTINPFDD461 pKa = 3.48ISITLVSDD469 pKa = 3.32TTFCRR474 pKa = 11.84EE475 pKa = 4.13DD476 pKa = 3.65FPPPFGTNGTASVEE490 pKa = 4.11AEE492 pKa = 3.69ISGTPTSVVWSNGQTGNILVPDD514 pKa = 3.75STGFYY519 pKa = 10.46YY520 pKa = 10.83VVATDD525 pKa = 3.6ATGCSTYY532 pKa = 11.16AGVQVNTYY540 pKa = 10.47GEE542 pKa = 3.78QDD544 pKa = 3.1QRR546 pKa = 11.84GNIWYY551 pKa = 9.16FGNNAGIDD559 pKa = 4.07FNPPPPTEE567 pKa = 4.98AIPFGDD573 pKa = 3.18ASIYY577 pKa = 10.94NGGNQLRR584 pKa = 11.84MPEE587 pKa = 4.01GCAIYY592 pKa = 10.06CDD594 pKa = 4.78RR595 pKa = 11.84NGQPLFYY602 pKa = 10.46SDD604 pKa = 3.74GVDD607 pKa = 3.55VFDD610 pKa = 6.22RR611 pKa = 11.84EE612 pKa = 4.62ANLLTPANKK621 pKa = 9.59IGGTAGISQGSSQSVLVLPFPGDD644 pKa = 3.23EE645 pKa = 4.01TLFYY649 pKa = 11.08LFTTNEE655 pKa = 4.0VYY657 pKa = 10.74NPSGSYY663 pKa = 8.19EE664 pKa = 3.75LRR666 pKa = 11.84YY667 pKa = 10.3SVFDD671 pKa = 3.94LKK673 pKa = 11.31LNNGLGGMVLDD684 pKa = 4.14QNGEE688 pKa = 3.68ISTILCSDD696 pKa = 3.21VTEE699 pKa = 4.04RR700 pKa = 11.84LAGNEE705 pKa = 3.39NWVIVHH711 pKa = 6.39EE712 pKa = 4.5LGNNNFRR719 pKa = 11.84AYY721 pKa = 9.39PVLGDD726 pKa = 4.21GIGTPVISNIGAIHH740 pKa = 6.04NSEE743 pKa = 4.23LEE745 pKa = 3.87SRR747 pKa = 11.84GYY749 pKa = 9.65MKK751 pKa = 11.07LNGNSNQLAVAYY763 pKa = 10.59SEE765 pKa = 4.27STNNNFVEE773 pKa = 5.98LFDD776 pKa = 4.64FDD778 pKa = 3.97NATGEE783 pKa = 4.24VSNYY787 pKa = 10.01LQLDD791 pKa = 3.87FNNPVSSAGPVSGQVYY807 pKa = 10.37GIEE810 pKa = 5.12FGNNDD815 pKa = 3.31NNLFASLIGTPSEE828 pKa = 3.72IFLWHH833 pKa = 6.72VDD835 pKa = 3.13TTTIAGNITDD845 pKa = 3.66PTYY848 pKa = 10.69IRR850 pKa = 11.84DD851 pKa = 3.66SVEE854 pKa = 4.12VVDD857 pKa = 6.22LSSLPATDD865 pKa = 5.2SIGALQLGPDD875 pKa = 3.56GQMYY879 pKa = 10.2GAIQNQQFTVTIGNTGTRR897 pKa = 11.84FGQPVPVANTFGLAAGTSSQLGLPNFIQFSGSSSQNASLAVEE939 pKa = 4.82SGCIDD944 pKa = 3.31QPLTFTVNNPLNPNLEE960 pKa = 4.15IYY962 pKa = 9.97FVSIFPEE969 pKa = 4.4GDD971 pKa = 3.07EE972 pKa = 4.68SNVLVGPIQLTQDD985 pKa = 3.35NPDD988 pKa = 3.45FTFSLPDD995 pKa = 3.75PGNYY999 pKa = 8.74VARR1002 pKa = 11.84FFIDD1006 pKa = 3.34QQCIPQQLITDD1017 pKa = 4.14SDD1019 pKa = 3.71QLFAINPLPTVSNIAVTDD1037 pKa = 3.69PTGCGLPDD1045 pKa = 3.83GSAIVTFSTTGNITYY1060 pKa = 10.07SVSGPVSYY1068 pKa = 10.57PDD1070 pKa = 3.18QTVTTTGSTNVTIPALSAGFYY1091 pKa = 9.42TLNAAFDD1098 pKa = 4.12TGCSEE1103 pKa = 4.21TFTFILNDD1111 pKa = 3.66PVPYY1115 pKa = 9.86TLAAVQSQGSDD1126 pKa = 3.21CDD1128 pKa = 3.89DD1129 pKa = 3.44QNGEE1133 pKa = 4.03ITVTLSPLAQVPAAYY1148 pKa = 9.96DD1149 pKa = 3.24WVLNEE1154 pKa = 4.02QSTGTTVRR1162 pKa = 11.84TGNQADD1168 pKa = 4.51DD1169 pKa = 4.39PDD1171 pKa = 4.71SDD1173 pKa = 4.12GVFVITGVSTGNYY1186 pKa = 9.49FLEE1189 pKa = 4.53LTDD1192 pKa = 3.63NAGCISTVNVSITPPPSITLTLPDD1216 pKa = 4.44QFFQACDD1223 pKa = 3.1VDD1225 pKa = 4.96EE1226 pKa = 4.31IVIPITTNSNSLVEE1240 pKa = 4.77LLSQTGDD1247 pKa = 3.19LLTNFRR1253 pKa = 11.84VIGQNDD1259 pKa = 4.15SIAITNPGISGEE1271 pKa = 4.09PIIFGIRR1278 pKa = 11.84ALGEE1282 pKa = 4.21DD1283 pKa = 5.1PISCDD1288 pKa = 3.09TTVVIQVIFDD1298 pKa = 3.62NSSPNPFDD1306 pKa = 4.31SRR1308 pKa = 11.84YY1309 pKa = 7.39TLCPFEE1315 pKa = 4.76SFEE1318 pKa = 4.29SGRR1321 pKa = 11.84QVVMLEE1327 pKa = 4.08NPPSGFQSARR1337 pKa = 11.84WFDD1340 pKa = 3.87RR1341 pKa = 11.84NSVEE1345 pKa = 4.29IEE1347 pKa = 4.09RR1348 pKa = 11.84INASGASNVTGYY1360 pKa = 10.62RR1361 pKa = 11.84FSQAGDD1367 pKa = 3.68SIIIEE1372 pKa = 3.99IAQPISVEE1380 pKa = 3.82LTNAFGCPTVASIDD1394 pKa = 3.39IVQDD1398 pKa = 3.19CKK1400 pKa = 11.31GRR1402 pKa = 11.84INAPSAFYY1410 pKa = 10.22PNSLIPRR1417 pKa = 11.84NRR1419 pKa = 11.84QFVVFPFLILPDD1431 pKa = 4.31DD1432 pKa = 3.73FQIYY1436 pKa = 9.96IFNRR1440 pKa = 11.84WGEE1443 pKa = 4.14MVFQSDD1449 pKa = 3.26NLQFMRR1455 pKa = 11.84EE1456 pKa = 4.01QGWNGGYY1463 pKa = 10.7DD1464 pKa = 3.84NDD1466 pKa = 4.66PGRR1469 pKa = 11.84PVPGGGYY1476 pKa = 10.37AYY1478 pKa = 10.23RR1479 pKa = 11.84INYY1482 pKa = 8.73KK1483 pKa = 9.78SQFEE1487 pKa = 3.92PDD1489 pKa = 3.16EE1490 pKa = 4.71LKK1492 pKa = 10.47EE1493 pKa = 3.95QRR1495 pKa = 11.84GGVTLIRR1502 pKa = 4.51
Molecular weight: 161.61 kDa
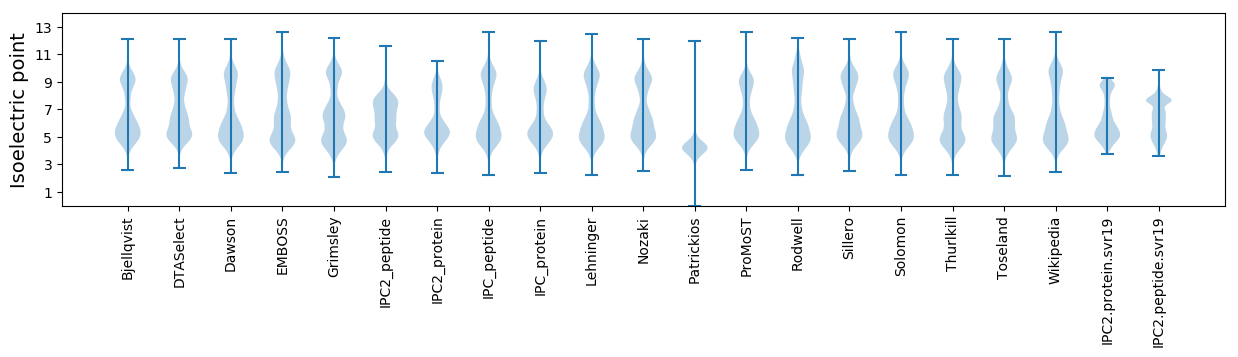
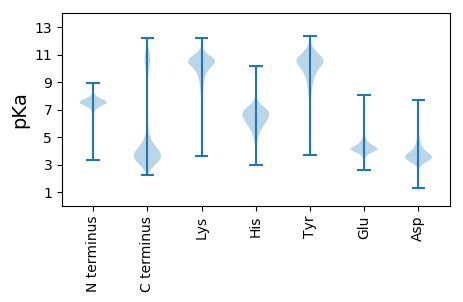
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A553FN91|A0A553FN91_9BACT Uncharacterized protein OS=Fulvivirga sp. M361 OX=2594266 GN=FNH22_12595 PE=4 SV=1
MM1 pKa = 7.88SSFTPWRR8 pKa = 11.84AYY10 pKa = 10.51SDD12 pKa = 3.26SAMRR16 pKa = 11.84ADD18 pKa = 3.99LGIGFVIRR26 pKa = 11.84VYY28 pKa = 10.76FRR30 pKa = 11.84GSFRR34 pKa = 11.84GMIRR38 pKa = 11.84QCFIIKK44 pKa = 8.93TIQNKK49 pKa = 9.58FIKK52 pKa = 10.36NMRR55 pKa = 11.84ITNAKK60 pKa = 10.1CSILKK65 pKa = 10.39EE66 pKa = 3.97NGNALRR72 pKa = 11.84FAKK75 pKa = 9.86YY76 pKa = 10.2APEE79 pKa = 4.58GDD81 pKa = 3.06IRR83 pKa = 11.84NIHH86 pKa = 7.02LSPQIEE92 pKa = 3.89NAAVNEE98 pKa = 3.86ARR100 pKa = 11.84ADD102 pKa = 3.57ALSRR106 pKa = 11.84LL107 pKa = 4.04
MM1 pKa = 7.88SSFTPWRR8 pKa = 11.84AYY10 pKa = 10.51SDD12 pKa = 3.26SAMRR16 pKa = 11.84ADD18 pKa = 3.99LGIGFVIRR26 pKa = 11.84VYY28 pKa = 10.76FRR30 pKa = 11.84GSFRR34 pKa = 11.84GMIRR38 pKa = 11.84QCFIIKK44 pKa = 8.93TIQNKK49 pKa = 9.58FIKK52 pKa = 10.36NMRR55 pKa = 11.84ITNAKK60 pKa = 10.1CSILKK65 pKa = 10.39EE66 pKa = 3.97NGNALRR72 pKa = 11.84FAKK75 pKa = 9.86YY76 pKa = 10.2APEE79 pKa = 4.58GDD81 pKa = 3.06IRR83 pKa = 11.84NIHH86 pKa = 7.02LSPQIEE92 pKa = 3.89NAAVNEE98 pKa = 3.86ARR100 pKa = 11.84ADD102 pKa = 3.57ALSRR106 pKa = 11.84LL107 pKa = 4.04
Molecular weight: 12.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2376036 |
24 |
4706 |
378.8 |
42.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.39 ± 0.025 | 0.736 ± 0.011 |
5.925 ± 0.024 | 6.539 ± 0.03 |
5.023 ± 0.022 | 6.892 ± 0.031 |
1.955 ± 0.016 | 7.119 ± 0.024 |
6.366 ± 0.039 | 9.44 ± 0.034 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.194 ± 0.015 | 5.504 ± 0.027 |
3.72 ± 0.019 | 3.567 ± 0.016 |
4.206 ± 0.019 | 6.831 ± 0.024 |
5.793 ± 0.034 | 6.451 ± 0.024 |
1.271 ± 0.013 | 4.079 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
