
Aquamicrobium defluvii
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Phyllobacteriaceae; Aquamicrobium
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins
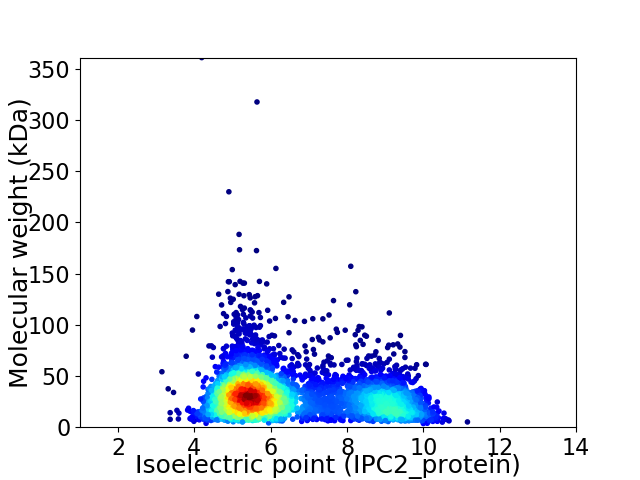
Virtual 2D-PAGE plot for 4454 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A011U0V1|A0A011U0V1_9RHIZ Uncharacterized protein OS=Aquamicrobium defluvii OX=69279 GN=BG36_14410 PE=4 SV=1
MM1 pKa = 7.83HH2 pKa = 7.4MDD4 pKa = 3.07STTEE8 pKa = 4.02IIAHH12 pKa = 6.5FIGVFSQTVEE22 pKa = 4.08DD23 pKa = 3.55VRR25 pKa = 11.84LRR27 pKa = 11.84LDD29 pKa = 3.42YY30 pKa = 11.42AKK32 pKa = 10.49FAAEE36 pKa = 3.95RR37 pKa = 11.84EE38 pKa = 4.47APPEE42 pKa = 4.15TGDD45 pKa = 4.49LLDD48 pKa = 4.59IKK50 pKa = 10.66LTMTSPYY57 pKa = 10.11TLGQFAPKK65 pKa = 8.6VTYY68 pKa = 10.27APPSFDD74 pKa = 2.88IVPTVAISFLSHH86 pKa = 6.0QPVPLHH92 pKa = 5.83MPPGNMTKK100 pKa = 10.12VFAAPHH106 pKa = 4.5QHH108 pKa = 6.09HH109 pKa = 6.52MPGMSEE115 pKa = 4.13RR116 pKa = 11.84MIPEE120 pKa = 3.93YY121 pKa = 10.88EE122 pKa = 4.66PIGSLALYY130 pKa = 9.84LRR132 pKa = 11.84QEE134 pKa = 3.88AHH136 pKa = 6.08LTDD139 pKa = 4.07NDD141 pKa = 3.73MLSIGDD147 pKa = 4.28HH148 pKa = 5.76GLAMKK153 pKa = 10.65AVGDD157 pKa = 4.05PVAALAKK164 pKa = 10.68LMGAASQLQPLADD177 pKa = 3.66MDD179 pKa = 4.34GLHH182 pKa = 6.29TEE184 pKa = 4.04ADD186 pKa = 3.06AGGFIQHH193 pKa = 5.47VTQAVDD199 pKa = 3.7TLPDD203 pKa = 3.7AAPAGGTLFVAQGAAAAGIHH223 pKa = 5.59VNGEE227 pKa = 4.38SVEE230 pKa = 4.1EE231 pKa = 4.13APVLEE236 pKa = 5.03DD237 pKa = 3.82LLPAKK242 pKa = 8.6FQPPAEE248 pKa = 4.28EE249 pKa = 3.96TSEE252 pKa = 5.48DD253 pKa = 3.83GDD255 pKa = 3.61GDD257 pKa = 4.02APVPTGVDD265 pKa = 3.45YY266 pKa = 11.32SLSSMIEE273 pKa = 3.86LEE275 pKa = 4.1AGGNLLVNEE284 pKa = 4.46AVLANNWYY292 pKa = 9.51GVPVVAAAGDD302 pKa = 4.07YY303 pKa = 9.79VHH305 pKa = 6.8VNAISQTNVWSDD317 pKa = 3.45CDD319 pKa = 3.7AVSDD323 pKa = 5.5ALGGWQRR330 pKa = 11.84LTDD333 pKa = 3.7TPTQAFNVATIEE345 pKa = 4.37TIANPAYY352 pKa = 10.39GEE354 pKa = 4.15AAGGAGFPSYY364 pKa = 9.67WQVSRR369 pKa = 11.84IDD371 pKa = 3.66GDD373 pKa = 4.22LLIANWIKK381 pKa = 10.4QVSFVQDD388 pKa = 2.82NDD390 pKa = 3.65VTVMSACGGGTTITLGDD407 pKa = 3.62NTVVNIASLAEE418 pKa = 3.96LGVYY422 pKa = 10.1YY423 pKa = 10.57DD424 pKa = 4.82LIIVGGSIYY433 pKa = 10.19HH434 pKa = 6.31GNIISQTNILLDD446 pKa = 4.59DD447 pKa = 4.85DD448 pKa = 5.21LVGTVGDD455 pKa = 4.05FQTSGEE461 pKa = 4.18ASLSTGGNLLWNQASITQYY480 pKa = 9.68GTMSFEE486 pKa = 4.15GLPPSFQAAADD497 pKa = 3.94TLASGPADD505 pKa = 3.75APDD508 pKa = 4.41DD509 pKa = 4.26LLHH512 pKa = 6.78HH513 pKa = 6.31EE514 pKa = 4.86AFGGLAGLKK523 pKa = 9.69VLYY526 pKa = 10.28ISGDD530 pKa = 3.85VIDD533 pKa = 5.02LQYY536 pKa = 11.04ISQTNILGDD545 pKa = 3.89ADD547 pKa = 3.98QVALAMDD554 pKa = 3.58QAGAYY559 pKa = 10.0ADD561 pKa = 4.53ADD563 pKa = 3.89WTVSTGTNALVNYY576 pKa = 9.51AHH578 pKa = 6.99IVDD581 pKa = 4.53GGVDD585 pKa = 3.47STVYY589 pKa = 10.78AGGDD593 pKa = 3.61VYY595 pKa = 11.33SDD597 pKa = 3.58EE598 pKa = 5.43LLIQAEE604 pKa = 4.8LISNDD609 pKa = 3.12SGLYY613 pKa = 8.5MQDD616 pKa = 2.11ATALVNEE623 pKa = 4.31AVVFLSDD630 pKa = 5.54DD631 pKa = 3.81LLAPDD636 pKa = 4.82AADD639 pKa = 4.52DD640 pKa = 4.17GAPVHH645 pKa = 7.16DD646 pKa = 4.62GAGPGAGSADD656 pKa = 3.52VMQTMLSS663 pKa = 3.51
MM1 pKa = 7.83HH2 pKa = 7.4MDD4 pKa = 3.07STTEE8 pKa = 4.02IIAHH12 pKa = 6.5FIGVFSQTVEE22 pKa = 4.08DD23 pKa = 3.55VRR25 pKa = 11.84LRR27 pKa = 11.84LDD29 pKa = 3.42YY30 pKa = 11.42AKK32 pKa = 10.49FAAEE36 pKa = 3.95RR37 pKa = 11.84EE38 pKa = 4.47APPEE42 pKa = 4.15TGDD45 pKa = 4.49LLDD48 pKa = 4.59IKK50 pKa = 10.66LTMTSPYY57 pKa = 10.11TLGQFAPKK65 pKa = 8.6VTYY68 pKa = 10.27APPSFDD74 pKa = 2.88IVPTVAISFLSHH86 pKa = 6.0QPVPLHH92 pKa = 5.83MPPGNMTKK100 pKa = 10.12VFAAPHH106 pKa = 4.5QHH108 pKa = 6.09HH109 pKa = 6.52MPGMSEE115 pKa = 4.13RR116 pKa = 11.84MIPEE120 pKa = 3.93YY121 pKa = 10.88EE122 pKa = 4.66PIGSLALYY130 pKa = 9.84LRR132 pKa = 11.84QEE134 pKa = 3.88AHH136 pKa = 6.08LTDD139 pKa = 4.07NDD141 pKa = 3.73MLSIGDD147 pKa = 4.28HH148 pKa = 5.76GLAMKK153 pKa = 10.65AVGDD157 pKa = 4.05PVAALAKK164 pKa = 10.68LMGAASQLQPLADD177 pKa = 3.66MDD179 pKa = 4.34GLHH182 pKa = 6.29TEE184 pKa = 4.04ADD186 pKa = 3.06AGGFIQHH193 pKa = 5.47VTQAVDD199 pKa = 3.7TLPDD203 pKa = 3.7AAPAGGTLFVAQGAAAAGIHH223 pKa = 5.59VNGEE227 pKa = 4.38SVEE230 pKa = 4.1EE231 pKa = 4.13APVLEE236 pKa = 5.03DD237 pKa = 3.82LLPAKK242 pKa = 8.6FQPPAEE248 pKa = 4.28EE249 pKa = 3.96TSEE252 pKa = 5.48DD253 pKa = 3.83GDD255 pKa = 3.61GDD257 pKa = 4.02APVPTGVDD265 pKa = 3.45YY266 pKa = 11.32SLSSMIEE273 pKa = 3.86LEE275 pKa = 4.1AGGNLLVNEE284 pKa = 4.46AVLANNWYY292 pKa = 9.51GVPVVAAAGDD302 pKa = 4.07YY303 pKa = 9.79VHH305 pKa = 6.8VNAISQTNVWSDD317 pKa = 3.45CDD319 pKa = 3.7AVSDD323 pKa = 5.5ALGGWQRR330 pKa = 11.84LTDD333 pKa = 3.7TPTQAFNVATIEE345 pKa = 4.37TIANPAYY352 pKa = 10.39GEE354 pKa = 4.15AAGGAGFPSYY364 pKa = 9.67WQVSRR369 pKa = 11.84IDD371 pKa = 3.66GDD373 pKa = 4.22LLIANWIKK381 pKa = 10.4QVSFVQDD388 pKa = 2.82NDD390 pKa = 3.65VTVMSACGGGTTITLGDD407 pKa = 3.62NTVVNIASLAEE418 pKa = 3.96LGVYY422 pKa = 10.1YY423 pKa = 10.57DD424 pKa = 4.82LIIVGGSIYY433 pKa = 10.19HH434 pKa = 6.31GNIISQTNILLDD446 pKa = 4.59DD447 pKa = 4.85DD448 pKa = 5.21LVGTVGDD455 pKa = 4.05FQTSGEE461 pKa = 4.18ASLSTGGNLLWNQASITQYY480 pKa = 9.68GTMSFEE486 pKa = 4.15GLPPSFQAAADD497 pKa = 3.94TLASGPADD505 pKa = 3.75APDD508 pKa = 4.41DD509 pKa = 4.26LLHH512 pKa = 6.78HH513 pKa = 6.31EE514 pKa = 4.86AFGGLAGLKK523 pKa = 9.69VLYY526 pKa = 10.28ISGDD530 pKa = 3.85VIDD533 pKa = 5.02LQYY536 pKa = 11.04ISQTNILGDD545 pKa = 3.89ADD547 pKa = 3.98QVALAMDD554 pKa = 3.58QAGAYY559 pKa = 10.0ADD561 pKa = 4.53ADD563 pKa = 3.89WTVSTGTNALVNYY576 pKa = 9.51AHH578 pKa = 6.99IVDD581 pKa = 4.53GGVDD585 pKa = 3.47STVYY589 pKa = 10.78AGGDD593 pKa = 3.61VYY595 pKa = 11.33SDD597 pKa = 3.58EE598 pKa = 5.43LLIQAEE604 pKa = 4.8LISNDD609 pKa = 3.12SGLYY613 pKa = 8.5MQDD616 pKa = 2.11ATALVNEE623 pKa = 4.31AVVFLSDD630 pKa = 5.54DD631 pKa = 3.81LLAPDD636 pKa = 4.82AADD639 pKa = 4.52DD640 pKa = 4.17GAPVHH645 pKa = 7.16DD646 pKa = 4.62GAGPGAGSADD656 pKa = 3.52VMQTMLSS663 pKa = 3.51
Molecular weight: 69.25 kDa
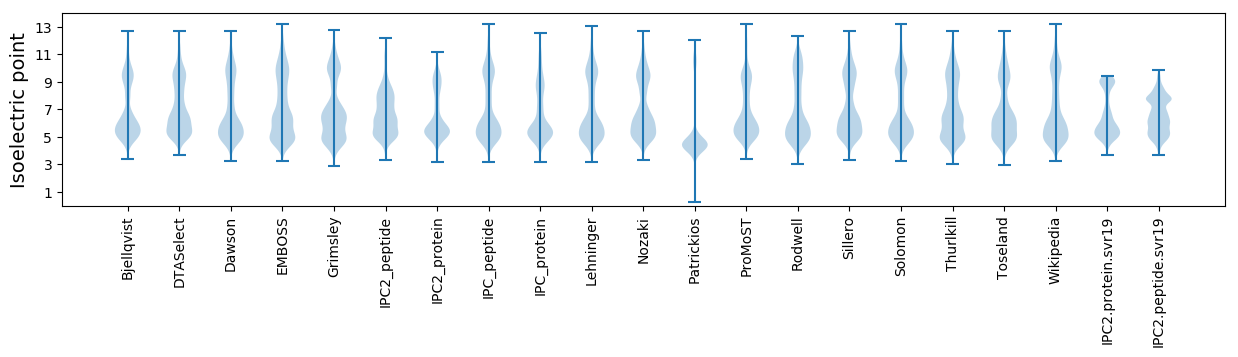
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A011UD08|A0A011UD08_9RHIZ Arsenic transporter OS=Aquamicrobium defluvii OX=69279 GN=BG36_11185 PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATNGGRR28 pKa = 11.84AVIAARR34 pKa = 11.84RR35 pKa = 11.84NRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATNGGRR28 pKa = 11.84AVIAARR34 pKa = 11.84RR35 pKa = 11.84NRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1348077 |
30 |
3895 |
302.7 |
32.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.404 ± 0.051 | 0.812 ± 0.011 |
5.676 ± 0.032 | 5.94 ± 0.038 |
3.763 ± 0.022 | 8.731 ± 0.073 |
2.039 ± 0.017 | 5.356 ± 0.026 |
3.223 ± 0.03 | 9.886 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.624 ± 0.019 | 2.593 ± 0.02 |
5.002 ± 0.031 | 3.054 ± 0.019 |
7.397 ± 0.045 | 5.442 ± 0.028 |
5.097 ± 0.021 | 7.453 ± 0.027 |
1.298 ± 0.012 | 2.211 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
