
Miniimonas arenae
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Beutenbergiaceae; Miniimonas
Average proteome isoelectric point is 6.03
Get precalculated fractions of proteins
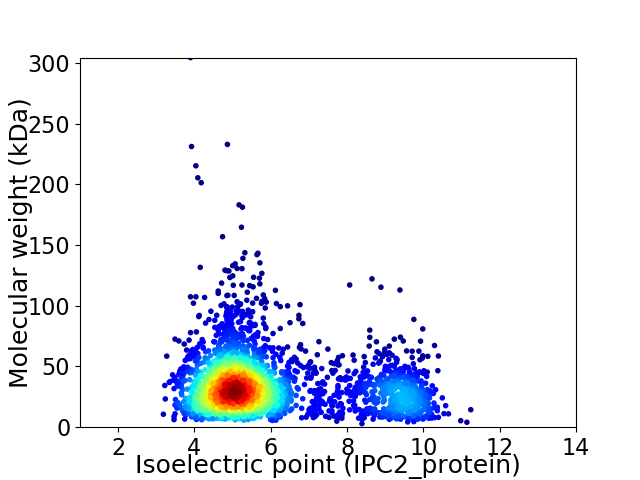
Virtual 2D-PAGE plot for 2846 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5C5BG52|A0A5C5BG52_9MICO UDP-N-acetylmuramyl-tripeptide synthetase OS=Miniimonas arenae OX=676201 GN=murE PE=3 SV=1
MM1 pKa = 7.73RR2 pKa = 11.84KK3 pKa = 9.48SLLSVAAVASVTLLLTACGGGGGGTDD29 pKa = 3.18ATGDD33 pKa = 3.63STATTDD39 pKa = 3.74ASAEE43 pKa = 4.05ATGDD47 pKa = 3.4AGASADD53 pKa = 3.64PSAPPVRR60 pKa = 11.84GNEE63 pKa = 4.08DD64 pKa = 3.54LVVWTDD70 pKa = 3.22ATKK73 pKa = 10.88LDD75 pKa = 3.76AVTAVAQAFGEE86 pKa = 4.38ANGITVGVQAIVNTRR101 pKa = 11.84SDD103 pKa = 4.58FITANQAGNGPDD115 pKa = 3.68VLVGAHH121 pKa = 6.58DD122 pKa = 4.43WIGQLVANGAIDD134 pKa = 4.08PLQISADD141 pKa = 3.79SLSGYY146 pKa = 7.39SEE148 pKa = 4.29KK149 pKa = 10.9AVTAVTYY156 pKa = 10.22NGQLMGIPYY165 pKa = 10.04GVEE168 pKa = 3.95SLVLYY173 pKa = 10.17CNKK176 pKa = 9.73TYY178 pKa = 11.02APDD181 pKa = 3.64TYY183 pKa = 10.94ATLDD187 pKa = 3.54DD188 pKa = 5.75AIAAGQAAKK197 pKa = 10.5DD198 pKa = 3.57AGSVQSALNIPQGVNGDD215 pKa = 4.13PYY217 pKa = 11.28HH218 pKa = 5.67MQPLFTSAGGYY229 pKa = 10.5LFGLDD234 pKa = 3.3ATGTPDD240 pKa = 4.71ANDD243 pKa = 3.86LGVGTDD249 pKa = 3.43AGIAAAQKK257 pKa = 10.18IATLGDD263 pKa = 3.58AGSKK267 pKa = 9.98VLSTAITNDD276 pKa = 3.01NAISLFAQGQAGCLISGPWALSDD299 pKa = 3.78VEE301 pKa = 4.94AGLGADD307 pKa = 4.83GYY309 pKa = 8.47TVQPIPGFAGMSPAEE324 pKa = 4.16PFMGAQAFYY333 pKa = 10.77VASNGQNKK341 pKa = 9.54SFAQAFVTGTSGGGLNSEE359 pKa = 4.59EE360 pKa = 4.3AMTTLFEE367 pKa = 4.42GTNLPPAMTSVRR379 pKa = 11.84ATAAASDD386 pKa = 3.94PLIGVFGDD394 pKa = 3.9AADD397 pKa = 3.76AAYY400 pKa = 9.63PMPAIPAMDD409 pKa = 3.8QVWTPLGQAYY419 pKa = 9.89AAIIGGADD427 pKa = 3.63PASTMTTAAQTISSAIASSS446 pKa = 3.24
MM1 pKa = 7.73RR2 pKa = 11.84KK3 pKa = 9.48SLLSVAAVASVTLLLTACGGGGGGTDD29 pKa = 3.18ATGDD33 pKa = 3.63STATTDD39 pKa = 3.74ASAEE43 pKa = 4.05ATGDD47 pKa = 3.4AGASADD53 pKa = 3.64PSAPPVRR60 pKa = 11.84GNEE63 pKa = 4.08DD64 pKa = 3.54LVVWTDD70 pKa = 3.22ATKK73 pKa = 10.88LDD75 pKa = 3.76AVTAVAQAFGEE86 pKa = 4.38ANGITVGVQAIVNTRR101 pKa = 11.84SDD103 pKa = 4.58FITANQAGNGPDD115 pKa = 3.68VLVGAHH121 pKa = 6.58DD122 pKa = 4.43WIGQLVANGAIDD134 pKa = 4.08PLQISADD141 pKa = 3.79SLSGYY146 pKa = 7.39SEE148 pKa = 4.29KK149 pKa = 10.9AVTAVTYY156 pKa = 10.22NGQLMGIPYY165 pKa = 10.04GVEE168 pKa = 3.95SLVLYY173 pKa = 10.17CNKK176 pKa = 9.73TYY178 pKa = 11.02APDD181 pKa = 3.64TYY183 pKa = 10.94ATLDD187 pKa = 3.54DD188 pKa = 5.75AIAAGQAAKK197 pKa = 10.5DD198 pKa = 3.57AGSVQSALNIPQGVNGDD215 pKa = 4.13PYY217 pKa = 11.28HH218 pKa = 5.67MQPLFTSAGGYY229 pKa = 10.5LFGLDD234 pKa = 3.3ATGTPDD240 pKa = 4.71ANDD243 pKa = 3.86LGVGTDD249 pKa = 3.43AGIAAAQKK257 pKa = 10.18IATLGDD263 pKa = 3.58AGSKK267 pKa = 9.98VLSTAITNDD276 pKa = 3.01NAISLFAQGQAGCLISGPWALSDD299 pKa = 3.78VEE301 pKa = 4.94AGLGADD307 pKa = 4.83GYY309 pKa = 8.47TVQPIPGFAGMSPAEE324 pKa = 4.16PFMGAQAFYY333 pKa = 10.77VASNGQNKK341 pKa = 9.54SFAQAFVTGTSGGGLNSEE359 pKa = 4.59EE360 pKa = 4.3AMTTLFEE367 pKa = 4.42GTNLPPAMTSVRR379 pKa = 11.84ATAAASDD386 pKa = 3.94PLIGVFGDD394 pKa = 3.9AADD397 pKa = 3.76AAYY400 pKa = 9.63PMPAIPAMDD409 pKa = 3.8QVWTPLGQAYY419 pKa = 9.89AAIIGGADD427 pKa = 3.63PASTMTTAAQTISSAIASSS446 pKa = 3.24
Molecular weight: 44.2 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5C5BDP5|A0A5C5BDP5_9MICO Prepilin peptidase OS=Miniimonas arenae OX=676201 GN=FH969_05510 PE=3 SV=1
MM1 pKa = 7.13GRR3 pKa = 11.84AGAGGSGRR11 pKa = 11.84AGVRR15 pKa = 11.84AGRR18 pKa = 11.84RR19 pKa = 11.84AGGRR23 pKa = 11.84GAGRR27 pKa = 11.84RR28 pKa = 11.84GRR30 pKa = 11.84GGQVRR35 pKa = 11.84TGRR38 pKa = 11.84GRR40 pKa = 11.84APTARR45 pKa = 11.84AANNGGGFGAIRR57 pKa = 11.84SEE59 pKa = 3.9TAAIVGRR66 pKa = 11.84RR67 pKa = 11.84RR68 pKa = 11.84TGARR72 pKa = 11.84SMLRR76 pKa = 11.84GRR78 pKa = 11.84FRR80 pKa = 11.84SAALRR85 pKa = 11.84KK86 pKa = 9.26RR87 pKa = 11.84PHH89 pKa = 6.08SARR92 pKa = 11.84ASYY95 pKa = 9.23GHH97 pKa = 6.9RR98 pKa = 11.84PHH100 pKa = 7.04ARR102 pKa = 11.84SQSRR106 pKa = 11.84RR107 pKa = 11.84AALGKK112 pKa = 10.03RR113 pKa = 11.84PRR115 pKa = 11.84KK116 pKa = 7.39RR117 pKa = 11.84TRR119 pKa = 11.84PATPRR124 pKa = 11.84RR125 pKa = 11.84TEE127 pKa = 3.53VRR129 pKa = 11.84RR130 pKa = 11.84SGRR133 pKa = 11.84TRR135 pKa = 3.0
MM1 pKa = 7.13GRR3 pKa = 11.84AGAGGSGRR11 pKa = 11.84AGVRR15 pKa = 11.84AGRR18 pKa = 11.84RR19 pKa = 11.84AGGRR23 pKa = 11.84GAGRR27 pKa = 11.84RR28 pKa = 11.84GRR30 pKa = 11.84GGQVRR35 pKa = 11.84TGRR38 pKa = 11.84GRR40 pKa = 11.84APTARR45 pKa = 11.84AANNGGGFGAIRR57 pKa = 11.84SEE59 pKa = 3.9TAAIVGRR66 pKa = 11.84RR67 pKa = 11.84RR68 pKa = 11.84TGARR72 pKa = 11.84SMLRR76 pKa = 11.84GRR78 pKa = 11.84FRR80 pKa = 11.84SAALRR85 pKa = 11.84KK86 pKa = 9.26RR87 pKa = 11.84PHH89 pKa = 6.08SARR92 pKa = 11.84ASYY95 pKa = 9.23GHH97 pKa = 6.9RR98 pKa = 11.84PHH100 pKa = 7.04ARR102 pKa = 11.84SQSRR106 pKa = 11.84RR107 pKa = 11.84AALGKK112 pKa = 10.03RR113 pKa = 11.84PRR115 pKa = 11.84KK116 pKa = 7.39RR117 pKa = 11.84TRR119 pKa = 11.84PATPRR124 pKa = 11.84RR125 pKa = 11.84TEE127 pKa = 3.53VRR129 pKa = 11.84RR130 pKa = 11.84SGRR133 pKa = 11.84TRR135 pKa = 3.0
Molecular weight: 14.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
974644 |
26 |
3032 |
342.5 |
36.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.834 ± 0.077 | 0.534 ± 0.011 |
6.298 ± 0.044 | 5.516 ± 0.046 |
2.475 ± 0.029 | 9.462 ± 0.043 |
2.078 ± 0.025 | 3.05 ± 0.039 |
1.313 ± 0.027 | 10.316 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.513 ± 0.021 | 1.471 ± 0.024 |
6.039 ± 0.04 | 2.551 ± 0.027 |
7.788 ± 0.061 | 5.157 ± 0.042 |
6.377 ± 0.053 | 9.904 ± 0.054 |
1.55 ± 0.021 | 1.774 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
