
Zhouia amylolytica AD3
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Zhouia; Zhouia amylolytica
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
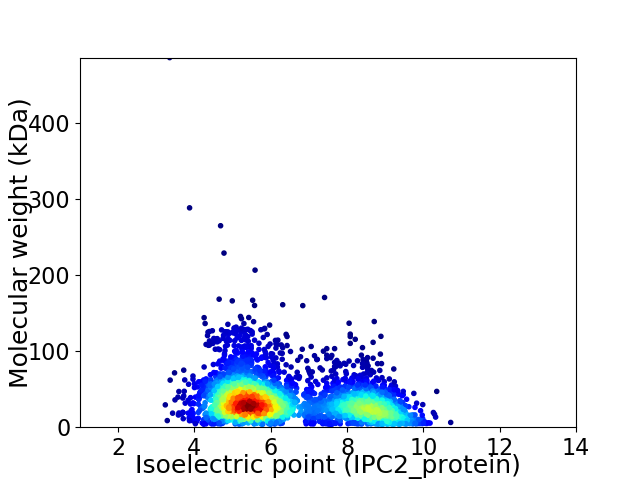
Virtual 2D-PAGE plot for 3353 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W2UL28|W2UL28_9FLAO Sulfate transporter OS=Zhouia amylolytica AD3 OX=1286632 GN=P278_26320 PE=3 SV=1
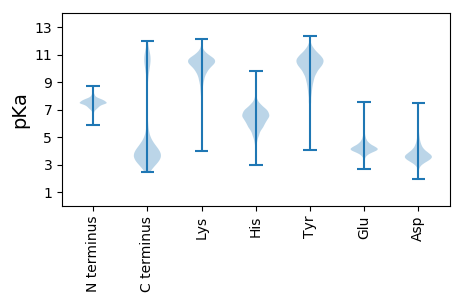
MM1 pKa = 7.64KK2 pKa = 10.22INYY5 pKa = 8.36KK6 pKa = 9.57WLLLLAFGFIACDD19 pKa = 3.46SDD21 pKa = 5.81DD22 pKa = 4.48DD23 pKa = 4.16MSSEE27 pKa = 4.21GPDD30 pKa = 3.05AVMLTSGEE38 pKa = 4.36ADD40 pKa = 3.44FSTYY44 pKa = 10.39VAVGSSLSAGYY55 pKa = 10.43SDD57 pKa = 3.8GALFIASQINSFPNILASKK76 pKa = 9.51FQMAGGGAFTQPLMGDD92 pKa = 3.46NLGGMLVGGNQLLEE106 pKa = 3.8NRR108 pKa = 11.84LFFNGEE114 pKa = 3.83LPEE117 pKa = 4.51RR118 pKa = 11.84LPGTPTTEE126 pKa = 4.16ATNVMPGPYY135 pKa = 9.92NNMGITGAKK144 pKa = 9.11IFHH147 pKa = 6.87LGVEE151 pKa = 5.48GYY153 pKa = 10.29GSSAALSVGAANPYY167 pKa = 7.2YY168 pKa = 11.07VRR170 pKa = 11.84MASSPGASVIGDD182 pKa = 4.2ALSLAPTFFTLWAGSDD198 pKa = 3.72DD199 pKa = 3.81VLSYY203 pKa = 10.91AVSGGIGEE211 pKa = 4.32DD212 pKa = 3.23QAGNFNPEE220 pKa = 4.12TYY222 pKa = 10.39GSNDD226 pKa = 2.75ITDD229 pKa = 3.92PAVFSQAFSGILTLLMSNGAKK250 pKa = 9.93GVVANIPDD258 pKa = 3.96VTALPYY264 pKa = 8.98FTTVPHH270 pKa = 6.92APLDD274 pKa = 3.73PTNEE278 pKa = 4.05DD279 pKa = 3.81FGPQIPLLNTIFGALNQIFVALGEE303 pKa = 4.32GEE305 pKa = 4.12RR306 pKa = 11.84AVVFSEE312 pKa = 4.73TSASPVVIKK321 pKa = 10.75DD322 pKa = 3.4EE323 pKa = 4.26SLTDD327 pKa = 3.36ISAQIEE333 pKa = 4.09AALNASEE340 pKa = 4.79TFPQFIAQFGLPPEE354 pKa = 4.51AAPGVANLLGLMYY367 pKa = 10.32GQSRR371 pKa = 11.84QATEE375 pKa = 3.78ADD377 pKa = 3.94LLVLTSSSVIGEE389 pKa = 4.25VNTAAVQFLISRR401 pKa = 11.84GLSQEE406 pKa = 3.84LAAQFSVEE414 pKa = 4.34GITYY418 pKa = 9.61PLDD421 pKa = 4.52DD422 pKa = 4.28GWVLLPSEE430 pKa = 4.1QNEE433 pKa = 4.2IKK435 pKa = 9.83TATTAFNTTIEE446 pKa = 4.36SLASQNGLAFVDD458 pKa = 4.04AHH460 pKa = 7.87ALMNEE465 pKa = 4.4VASGGLPFDD474 pKa = 3.94EE475 pKa = 4.84FTLNSSLVFGNTFSLDD491 pKa = 3.46GVHH494 pKa = 6.08PTARR498 pKa = 11.84GYY500 pKa = 11.57AFIANQFMLAIDD512 pKa = 3.86AAYY515 pKa = 10.16GSNFEE520 pKa = 4.25EE521 pKa = 4.89AGVLAKK527 pKa = 10.84AGDD530 pKa = 4.25YY531 pKa = 9.48GTMYY535 pKa = 10.4PVEE538 pKa = 4.31LPP540 pKa = 3.48
MM1 pKa = 7.64KK2 pKa = 10.22INYY5 pKa = 8.36KK6 pKa = 9.57WLLLLAFGFIACDD19 pKa = 3.46SDD21 pKa = 5.81DD22 pKa = 4.48DD23 pKa = 4.16MSSEE27 pKa = 4.21GPDD30 pKa = 3.05AVMLTSGEE38 pKa = 4.36ADD40 pKa = 3.44FSTYY44 pKa = 10.39VAVGSSLSAGYY55 pKa = 10.43SDD57 pKa = 3.8GALFIASQINSFPNILASKK76 pKa = 9.51FQMAGGGAFTQPLMGDD92 pKa = 3.46NLGGMLVGGNQLLEE106 pKa = 3.8NRR108 pKa = 11.84LFFNGEE114 pKa = 3.83LPEE117 pKa = 4.51RR118 pKa = 11.84LPGTPTTEE126 pKa = 4.16ATNVMPGPYY135 pKa = 9.92NNMGITGAKK144 pKa = 9.11IFHH147 pKa = 6.87LGVEE151 pKa = 5.48GYY153 pKa = 10.29GSSAALSVGAANPYY167 pKa = 7.2YY168 pKa = 11.07VRR170 pKa = 11.84MASSPGASVIGDD182 pKa = 4.2ALSLAPTFFTLWAGSDD198 pKa = 3.72DD199 pKa = 3.81VLSYY203 pKa = 10.91AVSGGIGEE211 pKa = 4.32DD212 pKa = 3.23QAGNFNPEE220 pKa = 4.12TYY222 pKa = 10.39GSNDD226 pKa = 2.75ITDD229 pKa = 3.92PAVFSQAFSGILTLLMSNGAKK250 pKa = 9.93GVVANIPDD258 pKa = 3.96VTALPYY264 pKa = 8.98FTTVPHH270 pKa = 6.92APLDD274 pKa = 3.73PTNEE278 pKa = 4.05DD279 pKa = 3.81FGPQIPLLNTIFGALNQIFVALGEE303 pKa = 4.32GEE305 pKa = 4.12RR306 pKa = 11.84AVVFSEE312 pKa = 4.73TSASPVVIKK321 pKa = 10.75DD322 pKa = 3.4EE323 pKa = 4.26SLTDD327 pKa = 3.36ISAQIEE333 pKa = 4.09AALNASEE340 pKa = 4.79TFPQFIAQFGLPPEE354 pKa = 4.51AAPGVANLLGLMYY367 pKa = 10.32GQSRR371 pKa = 11.84QATEE375 pKa = 3.78ADD377 pKa = 3.94LLVLTSSSVIGEE389 pKa = 4.25VNTAAVQFLISRR401 pKa = 11.84GLSQEE406 pKa = 3.84LAAQFSVEE414 pKa = 4.34GITYY418 pKa = 9.61PLDD421 pKa = 4.52DD422 pKa = 4.28GWVLLPSEE430 pKa = 4.1QNEE433 pKa = 4.2IKK435 pKa = 9.83TATTAFNTTIEE446 pKa = 4.36SLASQNGLAFVDD458 pKa = 4.04AHH460 pKa = 7.87ALMNEE465 pKa = 4.4VASGGLPFDD474 pKa = 3.94EE475 pKa = 4.84FTLNSSLVFGNTFSLDD491 pKa = 3.46GVHH494 pKa = 6.08PTARR498 pKa = 11.84GYY500 pKa = 11.57AFIANQFMLAIDD512 pKa = 3.86AAYY515 pKa = 10.16GSNFEE520 pKa = 4.25EE521 pKa = 4.89AGVLAKK527 pKa = 10.84AGDD530 pKa = 4.25YY531 pKa = 9.48GTMYY535 pKa = 10.4PVEE538 pKa = 4.31LPP540 pKa = 3.48
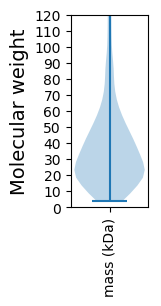
Molecular weight: 56.44 kDa
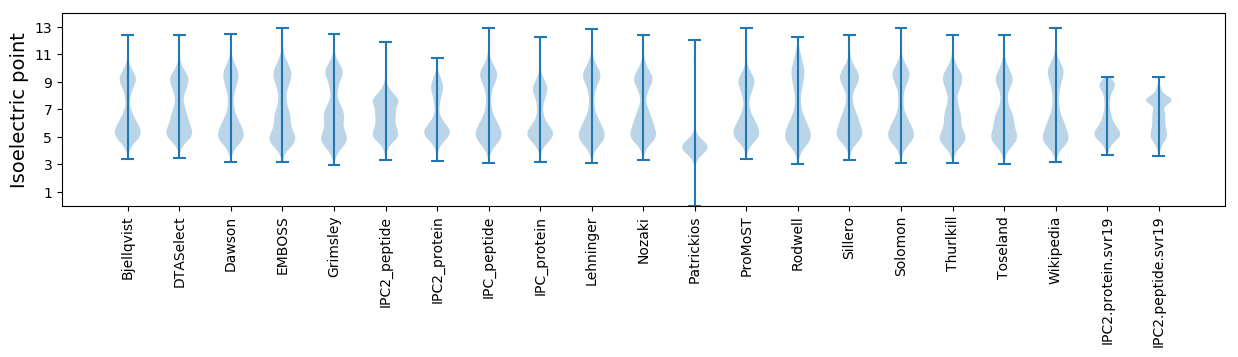
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W2UPN9|W2UPN9_9FLAO Uncharacterized protein OS=Zhouia amylolytica AD3 OX=1286632 GN=P278_16900 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 9.13RR10 pKa = 11.84KK11 pKa = 9.48RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.49HH16 pKa = 3.94GFRR19 pKa = 11.84EE20 pKa = 4.19RR21 pKa = 11.84MATANGRR28 pKa = 11.84KK29 pKa = 9.14VLAKK33 pKa = 10.18RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.07GRR39 pKa = 11.84KK40 pKa = 7.95KK41 pKa = 10.52LSVSSEE47 pKa = 3.87PRR49 pKa = 11.84HH50 pKa = 5.92KK51 pKa = 10.61KK52 pKa = 9.84
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 9.13RR10 pKa = 11.84KK11 pKa = 9.48RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.49HH16 pKa = 3.94GFRR19 pKa = 11.84EE20 pKa = 4.19RR21 pKa = 11.84MATANGRR28 pKa = 11.84KK29 pKa = 9.14VLAKK33 pKa = 10.18RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.07GRR39 pKa = 11.84KK40 pKa = 7.95KK41 pKa = 10.52LSVSSEE47 pKa = 3.87PRR49 pKa = 11.84HH50 pKa = 5.92KK51 pKa = 10.61KK52 pKa = 9.84
Molecular weight: 6.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1128980 |
37 |
4606 |
336.7 |
38.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.365 ± 0.041 | 0.708 ± 0.014 |
5.739 ± 0.037 | 6.859 ± 0.043 |
5.015 ± 0.034 | 6.554 ± 0.046 |
1.901 ± 0.023 | 7.699 ± 0.041 |
7.649 ± 0.057 | 9.204 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.287 ± 0.017 | 6.006 ± 0.032 |
3.511 ± 0.023 | 3.329 ± 0.024 |
3.595 ± 0.027 | 6.347 ± 0.033 |
5.496 ± 0.049 | 6.313 ± 0.034 |
1.15 ± 0.018 | 4.275 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
