
Rhinolophus ferrumequinum papillomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Treisdeltapapillomavirus; Treisdeltapapillomavirus 1
Average proteome isoelectric point is 5.99
Get precalculated fractions of proteins
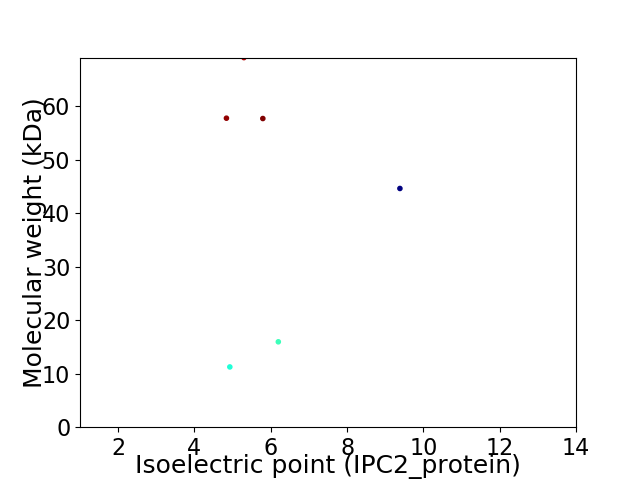
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
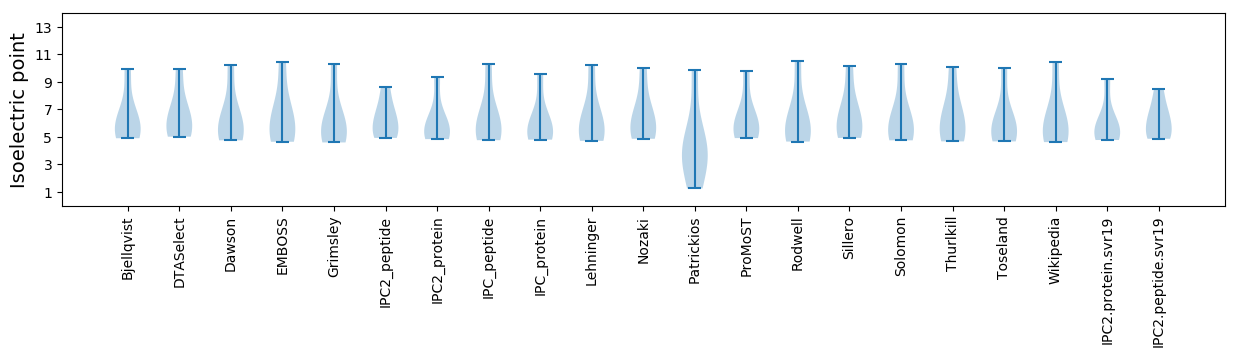
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W8EH64|W8EH64_9PAPI Protein E7 OS=Rhinolophus ferrumequinum papillomavirus 1 OX=1464074 GN=E7 PE=3 SV=1
MM1 pKa = 7.64RR2 pKa = 11.84GSHH5 pKa = 6.57IDD7 pKa = 3.36LRR9 pKa = 11.84DD10 pKa = 3.71IILQEE15 pKa = 3.95EE16 pKa = 4.81PEE18 pKa = 4.59NIDD21 pKa = 3.77LRR23 pKa = 11.84CDD25 pKa = 3.55EE26 pKa = 4.38ILQEE30 pKa = 4.28EE31 pKa = 4.77EE32 pKa = 4.68EE33 pKa = 4.33EE34 pKa = 4.32QQVHH38 pKa = 5.64GLISRR43 pKa = 11.84QAFQVAICCGLCQRR57 pKa = 11.84PIKK60 pKa = 9.62FVCLTTILALRR71 pKa = 11.84QLEE74 pKa = 4.03QLLFGPLDD82 pKa = 3.96FVCVRR87 pKa = 11.84CVNTHH92 pKa = 5.62EE93 pKa = 4.25LHH95 pKa = 7.14HH96 pKa = 6.64GGG98 pKa = 4.14
MM1 pKa = 7.64RR2 pKa = 11.84GSHH5 pKa = 6.57IDD7 pKa = 3.36LRR9 pKa = 11.84DD10 pKa = 3.71IILQEE15 pKa = 3.95EE16 pKa = 4.81PEE18 pKa = 4.59NIDD21 pKa = 3.77LRR23 pKa = 11.84CDD25 pKa = 3.55EE26 pKa = 4.38ILQEE30 pKa = 4.28EE31 pKa = 4.77EE32 pKa = 4.68EE33 pKa = 4.33EE34 pKa = 4.32QQVHH38 pKa = 5.64GLISRR43 pKa = 11.84QAFQVAICCGLCQRR57 pKa = 11.84PIKK60 pKa = 9.62FVCLTTILALRR71 pKa = 11.84QLEE74 pKa = 4.03QLLFGPLDD82 pKa = 3.96FVCVRR87 pKa = 11.84CVNTHH92 pKa = 5.62EE93 pKa = 4.25LHH95 pKa = 7.14HH96 pKa = 6.64GGG98 pKa = 4.14
Molecular weight: 11.27 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W8EG07|W8EG07_9PAPI Replication protein E1 OS=Rhinolophus ferrumequinum papillomavirus 1 OX=1464074 GN=E1 PE=3 SV=1
MM1 pKa = 7.06EE2 pKa = 4.73TLSSRR7 pKa = 11.84LEE9 pKa = 4.21SVQDD13 pKa = 3.75ALLTLYY19 pKa = 10.25EE20 pKa = 4.04QDD22 pKa = 3.89SNRR25 pKa = 11.84LDD27 pKa = 4.03DD28 pKa = 6.08QIKK31 pKa = 9.27QWTLVKK37 pKa = 10.55EE38 pKa = 4.4EE39 pKa = 4.12NVLLHH44 pKa = 6.81AARR47 pKa = 11.84KK48 pKa = 9.77KK49 pKa = 10.7GITRR53 pKa = 11.84LGMHH57 pKa = 6.62MVPTLQSSEE66 pKa = 3.55ARR68 pKa = 11.84ARR70 pKa = 11.84DD71 pKa = 3.99AIEE74 pKa = 3.74MEE76 pKa = 5.03LYY78 pKa = 10.57LKK80 pKa = 10.02SLRR83 pKa = 11.84QSPYY87 pKa = 9.19GTEE90 pKa = 3.86PWNLAQTTRR99 pKa = 11.84EE100 pKa = 4.04RR101 pKa = 11.84FLAAPPYY108 pKa = 9.95CFKK111 pKa = 11.01KK112 pKa = 10.6DD113 pKa = 3.48GSNVDD118 pKa = 3.19VYY120 pKa = 11.83YY121 pKa = 11.1DD122 pKa = 3.59NDD124 pKa = 3.43KK125 pKa = 11.3DD126 pKa = 3.72NSVRR130 pKa = 11.84YY131 pKa = 7.88ITWGAIYY138 pKa = 9.38YY139 pKa = 10.46QNGNDD144 pKa = 4.03DD145 pKa = 3.13WHH147 pKa = 6.43KK148 pKa = 8.04VQGRR152 pKa = 11.84IEE154 pKa = 4.42HH155 pKa = 7.01DD156 pKa = 2.96GLYY159 pKa = 10.06YY160 pKa = 10.21VQHH163 pKa = 7.46DD164 pKa = 4.43GLKK167 pKa = 8.52VTYY170 pKa = 10.66VDD172 pKa = 5.29FAAEE176 pKa = 3.7ASAYY180 pKa = 10.52SRR182 pKa = 11.84TGSWRR187 pKa = 11.84VVYY190 pKa = 10.47NNKK193 pKa = 8.32TVLPDD198 pKa = 3.29NSVASSRR205 pKa = 11.84SGRR208 pKa = 11.84QRR210 pKa = 11.84TSSPTGPGAEE220 pKa = 3.95YY221 pKa = 10.46SPRR224 pKa = 11.84STTPTDD230 pKa = 3.47TVVATPLKK238 pKa = 10.52ASKK241 pKa = 10.3KK242 pKa = 8.44DD243 pKa = 3.13TAAGVRR249 pKa = 11.84RR250 pKa = 11.84GTRR253 pKa = 11.84HH254 pKa = 5.78HH255 pKa = 6.7NGAGRR260 pKa = 11.84RR261 pKa = 11.84GGGRR265 pKa = 11.84RR266 pKa = 11.84VQQRR270 pKa = 11.84KK271 pKa = 9.42SDD273 pKa = 3.97PKK275 pKa = 9.98RR276 pKa = 11.84GAAITPPSPSEE287 pKa = 3.61VGGRR291 pKa = 11.84HH292 pKa = 4.37QTPQRR297 pKa = 11.84RR298 pKa = 11.84SGSRR302 pKa = 11.84LQRR305 pKa = 11.84LLQEE309 pKa = 4.8ARR311 pKa = 11.84DD312 pKa = 3.97PPAVLLKK319 pKa = 10.81GQANTLKK326 pKa = 10.56CFRR329 pKa = 11.84FQAKK333 pKa = 10.07AKK335 pKa = 10.06FAGQFRR341 pKa = 11.84SVSTTFYY348 pKa = 7.95WTASEE353 pKa = 4.2GTDD356 pKa = 2.73RR357 pKa = 11.84VGRR360 pKa = 11.84ARR362 pKa = 11.84VIFLFTDD369 pKa = 3.24KK370 pKa = 10.95GQRR373 pKa = 11.84EE374 pKa = 4.11RR375 pKa = 11.84FMSRR379 pKa = 11.84VKK381 pKa = 10.38FPPSIEE387 pKa = 4.2TVLLNMDD394 pKa = 4.28GFF396 pKa = 4.35
MM1 pKa = 7.06EE2 pKa = 4.73TLSSRR7 pKa = 11.84LEE9 pKa = 4.21SVQDD13 pKa = 3.75ALLTLYY19 pKa = 10.25EE20 pKa = 4.04QDD22 pKa = 3.89SNRR25 pKa = 11.84LDD27 pKa = 4.03DD28 pKa = 6.08QIKK31 pKa = 9.27QWTLVKK37 pKa = 10.55EE38 pKa = 4.4EE39 pKa = 4.12NVLLHH44 pKa = 6.81AARR47 pKa = 11.84KK48 pKa = 9.77KK49 pKa = 10.7GITRR53 pKa = 11.84LGMHH57 pKa = 6.62MVPTLQSSEE66 pKa = 3.55ARR68 pKa = 11.84ARR70 pKa = 11.84DD71 pKa = 3.99AIEE74 pKa = 3.74MEE76 pKa = 5.03LYY78 pKa = 10.57LKK80 pKa = 10.02SLRR83 pKa = 11.84QSPYY87 pKa = 9.19GTEE90 pKa = 3.86PWNLAQTTRR99 pKa = 11.84EE100 pKa = 4.04RR101 pKa = 11.84FLAAPPYY108 pKa = 9.95CFKK111 pKa = 11.01KK112 pKa = 10.6DD113 pKa = 3.48GSNVDD118 pKa = 3.19VYY120 pKa = 11.83YY121 pKa = 11.1DD122 pKa = 3.59NDD124 pKa = 3.43KK125 pKa = 11.3DD126 pKa = 3.72NSVRR130 pKa = 11.84YY131 pKa = 7.88ITWGAIYY138 pKa = 9.38YY139 pKa = 10.46QNGNDD144 pKa = 4.03DD145 pKa = 3.13WHH147 pKa = 6.43KK148 pKa = 8.04VQGRR152 pKa = 11.84IEE154 pKa = 4.42HH155 pKa = 7.01DD156 pKa = 2.96GLYY159 pKa = 10.06YY160 pKa = 10.21VQHH163 pKa = 7.46DD164 pKa = 4.43GLKK167 pKa = 8.52VTYY170 pKa = 10.66VDD172 pKa = 5.29FAAEE176 pKa = 3.7ASAYY180 pKa = 10.52SRR182 pKa = 11.84TGSWRR187 pKa = 11.84VVYY190 pKa = 10.47NNKK193 pKa = 8.32TVLPDD198 pKa = 3.29NSVASSRR205 pKa = 11.84SGRR208 pKa = 11.84QRR210 pKa = 11.84TSSPTGPGAEE220 pKa = 3.95YY221 pKa = 10.46SPRR224 pKa = 11.84STTPTDD230 pKa = 3.47TVVATPLKK238 pKa = 10.52ASKK241 pKa = 10.3KK242 pKa = 8.44DD243 pKa = 3.13TAAGVRR249 pKa = 11.84RR250 pKa = 11.84GTRR253 pKa = 11.84HH254 pKa = 5.78HH255 pKa = 6.7NGAGRR260 pKa = 11.84RR261 pKa = 11.84GGGRR265 pKa = 11.84RR266 pKa = 11.84VQQRR270 pKa = 11.84KK271 pKa = 9.42SDD273 pKa = 3.97PKK275 pKa = 9.98RR276 pKa = 11.84GAAITPPSPSEE287 pKa = 3.61VGGRR291 pKa = 11.84HH292 pKa = 4.37QTPQRR297 pKa = 11.84RR298 pKa = 11.84SGSRR302 pKa = 11.84LQRR305 pKa = 11.84LLQEE309 pKa = 4.8ARR311 pKa = 11.84DD312 pKa = 3.97PPAVLLKK319 pKa = 10.81GQANTLKK326 pKa = 10.56CFRR329 pKa = 11.84FQAKK333 pKa = 10.07AKK335 pKa = 10.06FAGQFRR341 pKa = 11.84SVSTTFYY348 pKa = 7.95WTASEE353 pKa = 4.2GTDD356 pKa = 2.73RR357 pKa = 11.84VGRR360 pKa = 11.84ARR362 pKa = 11.84VIFLFTDD369 pKa = 3.24KK370 pKa = 10.95GQRR373 pKa = 11.84EE374 pKa = 4.11RR375 pKa = 11.84FMSRR379 pKa = 11.84VKK381 pKa = 10.38FPPSIEE387 pKa = 4.2TVLLNMDD394 pKa = 4.28GFF396 pKa = 4.35
Molecular weight: 44.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2282 |
98 |
610 |
380.3 |
42.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.179 ± 0.429 | 2.016 ± 0.758 |
6.31 ± 0.262 | 6.486 ± 0.646 |
4.689 ± 0.333 | 6.573 ± 0.665 |
2.235 ± 0.168 | 4.251 ± 0.558 |
4.514 ± 0.864 | 8.282 ± 0.802 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.972 ± 0.458 | 3.769 ± 0.586 |
6.223 ± 0.899 | 4.514 ± 0.417 |
6.836 ± 0.851 | 7.406 ± 0.7 |
6.398 ± 0.569 | 7.143 ± 0.729 |
1.227 ± 0.369 | 2.98 ± 0.348 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
